HFE
HFE is named after High Fe (iron), and unusually is not named after its associated disease hemochromatosis that led to the genes identification. The gene products known function is to regulate iron absorption by interacting with the transferrin receptor (TfR). Both HFE and transferrin (Tf) compete with each other for binding to the receptor (TfR)[1]. Tf binds to iron in the form of diferric transferrin (Fe-Tf or holo-Tf[2]) and in turn binds to free TfR which imports the iron into the cell by an endocytosis cycle[3]. The peptide hormone hepcidin is produced in the liver, increased hepcidin levels inhibit iron uptake by degrading the ferroportin (Fpn) iron exporter to the blood. Unbound HFE in the presence of holo-Tf also induces higher transcription levels of hepcidin via an unclear mechanism[4]. When the normal function of HFE is disrupted by mutation the result can be HFE hereditary hemochromatosis or toxic iron overload.
OMIM: 613609[1] NCBI Gene: 3077[2]
DNA Sequence
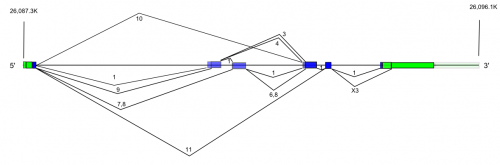
HFE is located on chromosome 6 in humans, in 7 exons over 12 kbp.
Cytogenetic location: 6p22.2
Genomic coordinates (GRCh37): 6:26,087,421 - 26,096,437
RNA Sequence
There are several alternatively spliced variants of HFE.
>gi|91718876|ref|NM_000410.3| Homo sapiens hemochromatosis (HFE), transcript variant 1, mRNA CTAAAGTTCTGAAAGACCTGTTGCTTTTCACCAGGAAGTTTTACTGGGCATCTCCTGAGCCTAGGCAATA GCTGTAGGGTGACTTCTGGAGCCATCCCCGTTTCCCCGCCCCCCAAAAGAAGCGGAGATTTAACGGGGAC GTGCGGCCAGAGCTGGGGAAATGGGCCCGCGAGCCAGGCCGGCGCTTCTCCTCCTGATGCTTTTGCAGAC CGCGGTCCTGCAGGGGCGCTTGCTGCGTTCACACTCTCTGCACTACCTCTTCATGGGTGCCTCAGAGCAG GACCTTGGTCTTTCCTTGTTTGAAGCTTTGGGCTACGTGGATGACCAGCTGTTCGTGTTCTATGATCATG AGAGTCGCCGTGTGGAGCCCCGAACTCCATGGGTTTCCAGTAGAATTTCAAGCCAGATGTGGCTGCAGCT GAGTCAGAGTCTGAAAGGGTGGGATCACATGTTCACTGTTGACTTCTGGACTATTATGGAAAATCACAAC CACAGCAAGGAGTCCCACACCCTGCAGGTCATCCTGGGCTGTGAAATGCAAGAAGACAACAGTACCGAGG GCTACTGGAAGTACGGGTATGATGGGCAGGACCACCTTGAATTCTGCCCTGACACACTGGATTGGAGAGC AGCAGAACCCAGGGCCTGGCCCACCAAGCTGGAGTGGGAAAGGCACAAGATTCGGGCCAGGCAGAACAGG GCCTACCTGGAGAGGGACTGCCCTGCACAGCTGCAGCAGTTGCTGGAGCTGGGGAGAGGTGTTTTGGACC AACAAGTGCCTCCTTTGGTGAAGGTGACACATCATGTGACCTCTTCAGTGACCACTCTACGGTGTCGGGC CTTGAACTACTACCCCCAGAACATCACCATGAAGTGGCTGAAGGATAAGCAGCCAATGGATGCCAAGGAG TTCGAACCTAAAGACGTATTGCCCAATGGGGATGGGACCTACCAGGGCTGGATAACCTTGGCTGTACCCC CTGGGGAAGAGCAGAGATATACGTGCCAGGTGGAGCACCCAGGCCTGGATCAGCCCCTCATTGTGATCTG GGAGCCCTCACCGTCTGGCACCCTAGTCATTGGAGTCATCAGTGGAATTGCTGTTTTTGTCGTCATCTTG TTCATTGGAATTTTGTTCATAATATTAAGGAAGAGGCAGGGTTCAAGAGGAGCCATGGGGCACTACGTCT TAGCTGAACGTGAGTGACACGCAGCCTGCAGACTCACTGTGGGAAGGAGACAAAACTAGAGACTCAAAGA GGGAGTGCATTTATGAGCTCTTCATGTTTCAGGAGAGAGTTGAACCTAAACATAGAAATTGCCTGACGAA CTCCTTGATTTTAGCCTTCTCTGTTCATTTCCTCAAAAAGATTTCCCCATTTAGGTTTCTGAGTTCCTGC ATGCCGGTGATCCCTAGCTGTGACCTCTCCCCTGGAACTGTCTCTCATGAACCTCAAGCTGCATCTAGAG GCTTCCTTCATTTCCTCCGTCACCTCAGAGACATACACCTATGTCATTTCATTTCCTATTTTTGGAAGAG GACTCCTTAAATTTGGGGGACTTACATGATTCATTTTAACATCTGAGAAAAGCTTTGAACCCTGGGACGT GGCTAGTCATAACCTTACCAGATTTTTACACATGTATCTATGCATTTTCTGGACCCGTTCAACTTTTCCT TTGAATCCTCTCTCTGTGTTACCCAGTAACTCATCTGTCACCAAGCCTTGGGGATTCTTCCATCTGATTG TGATGTGAGTTGCACAGCTATGAAGGCTGTACACTGCACGAATGGAAGAGGCACCTGTCCCAGAAAAAGC ATCATGGCTATCTGTGGGTAGTATGATGGGTGTTTTTAGCAGGTAGGAGGCAAATATCTTGAAAGGGGTT GTGAAGAGGTGTTTTTTCTAATTGGCATGAAGGTGTCATACAGATTTGCAAAGTTTAATGGTGCCTTCAT TTGGGATGCTACTCTAGTATTCCAGACCTGAAGAATCACAATAATTTTCTACCTGGTCTCTCCTTGTTCT GATAATGAAAATTATGATAAGGATGATAAAAGCACTTACTTCGTGTCCGACTCTTCTGAGCACCTACTTA CATGCATTACTGCATGCACTTCTTACAATAATTCTATGAGATAGGTACTATTATCCCCATTTCTTTTTTA AATGAAGAAAGTGAAGTAGGCCGGGCACGGTGGCTCACGCCTGTAATCCCAG
Codons without UTRs
ATGGGCCCGCGAGCCAGGCCGGCGCTTCTCCTCCTGATGCTTTTGCAGACCGCGGTCCTGCAGGG GCGCTTGCTGCGTTCACACTCTCTGCACTACCTCTTCATGGGTGCCTCAGAGCAGGACCTTGGTCTTTCC TTGTTTGAAGCTTTGGGCTACGTGGATGACCAGCTGTTCGTGTTCTATGATCATGAGAGTCGCCGTGTGG AGCCCCGAACTCCATGGGTTTCCAGTAGAATTTCAAGCCAGATGTGGCTGCAGCTGAGTCAGAGTCTGAA AGGGTGGGATCACATGTTCACTGTTGACTTCTGGACTATTATGGAAAATCACAACCACAGCAAGGAGTCC CACACCCTGCAGGTCATCCTGGGCTGTGAAATGCAAGAAGACAACAGTACCGAGGGCTACTGGAAGTACG GGTATGATGGGCAGGACCACCTTGAATTCTGCCCTGACACACTGGATTGGAGAGCAGCAGAACCCAGGGC CTGGCCCACCAAGCTGGAGTGGGAAAGGCACAAGATTCGGGCCAGGCAGAACAGGGCCTACCTGGAGAGG GACTGCCCTGCACAGCTGCAGCAGTTGCTGGAGCTGGGGAGAGGTGTTTTGGACCAACAAGTGCCTCCTT TGGTGAAGGTGACACATCATGTGACCTCTTCAGTGACCACTCTACGGTGTCGGGCCTTGAACTACTACCC CCAGAACATCACCATGAAGTGGCTGAAGGATAAGCAGCCAATGGATGCCAAGGAGTTCGAACCTAAAGAC GTATTGCCCAATGGGGATGGGACCTACCAGGGCTGGATAACCTTGGCTGTACCCCCTGGGGAAGAGCAGA GATATACGTGCCAGGTGGAGCACCCAGGCCTGGATCAGCCCCTCATTGTGATCTGGGAGCCCTCACCGTC TGGCACCCTAGTCATTGGAGTCATCAGTGGAATTGCTGTTTTTGTCGTCATCTTGTTCATTGGAATTTTG TTCATAATATTAAGGAAGAGGCAGGGTTCAAGAGGAGCCATGGGGCACTACGTCTTAGCTGAACGTGAGT GA
Protein Sequence
The protein has a signal sequence and transmembrane domain. It forms a tertiary complex with beta2-microglobulin (beta2M) and TfR. HFE has sequence and structural similarity to MHC class I-type proteins.
Uniprot[3]
NCBI Protein[4]
>gi|4504377|ref|NP_000401.1| hereditary hemochromatosis protein isoform 1 precursor [Homo sapiens] MGPRARPALLLLMLLQTAVLQGRLLRSHSLHYLFMGASEQDLGLSLFEALGYVDDQLFVFYDHESRRVEP RTPWVSSRISSQMWLQLSQSLKGWDHMFTVDFWTIMENHNHSKESHTLQVILGCEMQEDNSTEGYWKYGY DGQDHLEFCPDTLDWRAAEPRAWPTKLEWERHKIRARQNRAYLERDCPAQLQQLLELGRGVLDQQVPPLV KVTHHVTSSVTTLRCRALNYYPQNITMKWLKDKQPMDAKEFEPKDVLPNGDGTYQGWITLAVPPGEEQRY TCQVEHPGLDQPLIVIWEPSPSGTLVIGVISGIAVFVVILFIGILFIILRKRQGSRGAMGHYVLAERE
NCBI Structure[5]
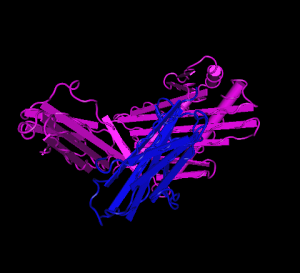
HFE (magenta) tertiary complex with beta2M (blue)[5]
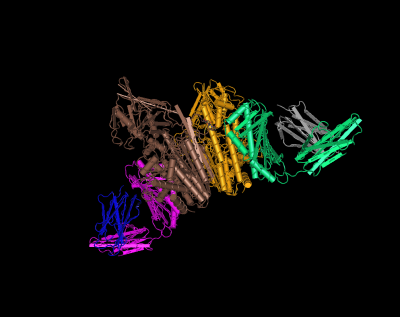
HFE (magenta and green) tertiary complex with beta2M (blue and gray) and TfR (yellow and brown)[6]
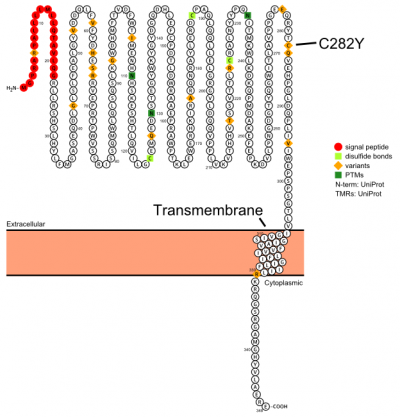
Plot in Protter[6] with UniProt accession: Q30201
To Do
Pelham, C., Jimenez, T., Rodova, M., Rudolph, A., Chipps, E., & Islam, M. R. (2013). Regulation of HFE expression by poly (ADP-ribose) polymerase-1 (PARP1) through an inverted repeat DNA sequence in the distal promoter. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 1829(12), 1257-1265.
Hentze, M. W., Muckenthaler, M. U., Galy, B., & Camaschella, C. (2010). Two to tango: regulation of Mammalian iron metabolism. Cell, 142(1), 24-38.
Wang, J., & Pantopoulos, K. (2011). Regulation of cellular iron metabolism. Biochem. J, 434, 365-381.
Fleming, R. E., & Ponka, P. (2012). Iron overload in human disease. New England Journal of Medicine, 366(4), 348-359.
Beutler, E., Felitti, V. J., Koziol, J. A., Ho, N. J., & Gelbart, T. (2002). Penetrance of 845G→ A (C282Y)< i> HFE</i> hereditary haemochromatosis mutation in the USA. The Lancet, 359(9302), 211-218.
Waheed, A., Parkkila, S., Zhou, X. Y., Tomatsu, S., Tsuchihashi, Z., Feder, J. N., ... & Sly, W. S. (1997). Hereditary hemochromatosis: effects of C282Y and H63D mutations on association with β2-microglobulin, intracellular processing, and cell surface expression of the HFE protein in COS-7 cells. Proceedings of the National Academy of Sciences, 94(23), 12384-12389.
Levy, J. E., Montross, L. K., Cohen, D. E., Fleming, M. D., & Andrews, N. C. (1999). The C282Y mutation causing hereditary hemochromatosis does not produce a null allele. Blood, 94(1), 9-11.
Waheed, A., Parkkila, S., Zhou, X. Y., Tomatsu, S., Tsuchihashi, Z., Feder, J. N., ... & Sly, W. S. (1997). Hereditary hemochromatosis: effects of C282Y and H63D mutations on association with β2-microglobulin, intracellular processing, and cell surface expression of the HFE protein in COS-7 cells. Proceedings of the National Academy of Sciences, 94(23), 12384-12389.
Mura, C., Raguenes, O., & Férec, C. (1999). HFE mutations analysis in 711 hemochromatosis probands: evidence for S65C implication in mild form of hemochromatosis. Blood, 93(8), 2502-2505.
Zhou, X. Y., Tomatsu, S., Fleming, R. E., Parkkila, S., Waheed, A., Jiang, J., ... & Sly, W. S. (1998). HFE gene knockout produces mouse model of hereditary hemochromatosis. Proceedings of the National Academy of Sciences, 95(5), 2492-2497.
Feder, J. N., Penny, D. M., Irrinki, A., Lee, V. K., Lebrón, J. A., Watson, N., ... & Schatzman, R. C. (1998). The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proceedings of the National Academy of Sciences, 95(4), 1472-1477.
Pietrangelo, A. (2004). Hereditary hemochromatosis—a new look at an old disease. New England Journal of Medicine, 350(23), 2383-2397.
Montosi, G., Donovan, A., Totaro, A., Garuti, C., Pignatti, E., Cassanelli, S., ... & Pietrangelo, A. (2001). Autosomal-dominant hemochromatosis is associated with a mutation in the ferroportin (SLC11A3) gene. Journal of Clinical Investigation, 108(4), 619-623.
Weiss, G. (2009). Genetic mechanisms and modifying factors in hereditary hemochromatosis. Nature Reviews Gastroenterology and Hepatology, 7(1), 50-58.
References
- ↑ Lebrón, J. A., West Jr, A. P., & Bjorkman, P. J. (1999). The hemochromatosis protein HFE competes with transferrin for binding to the transferrin receptor. Journal of molecular biology, 294(1), 239-245.
- ↑ The holo- prefix refers to a protein bound to its ligand. apo- is the unbound form of the protein.
- ↑ Qian, Z. M., Li, H., Sun, H., & Ho, K. (2002). Targeted drug delivery via the transferrin receptor-mediated endocytosis pathway. Pharmacological reviews, 54(4), 561-587.
- ↑ Gao, J., Chen, J., Kramer, M., Tsukamoto, H., Zhang, A. S., & Enns, C. A. (2009). Interaction of the hereditary hemochromatosis protein HFE with transferrin receptor 2 is required for transferrin-induced hepcidin expression. Cell metabolism, 9(3), 217-227.
- ↑ Lebron, J. A., Bennett, M. J., Vaughn, D. E., Chirino, A. J., Snow, P. M., Mintier, G. A., ... & Bjorkman, P. J. (1998). Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell, 93(1), 111-123.
- ↑ Bennett, M. J., Lebrón, J. A., & Bjorkman, P. J. (2000). Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature, 403(6765), 46-53.