Difference between revisions of "Begun and Aquadro 1992"
(→Notes) |
(→Notes) |
||
| (12 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | = | + | =Reference= |
Begun, D. J., & Aquadro, C. F. (1992). Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature, 356(6369), 519. | Begun, D. J., & Aquadro, C. F. (1992). Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature, 356(6369), 519. | ||
| Line 14: | Line 14: | ||
[[File:BegunAndAquadro1992Figures.png]] | [[File:BegunAndAquadro1992Figures.png]] | ||
| − | Under the neutral theory diversity within a species is expected to be | + | Under the neutral theory diversity within a species is expected to be 4''Nμ'' (''N'' is the population size) and differences between species is expected to be 2''tμ'' (''t'' is the time of divergence in generations). Both of these are proportional to the mutation rate ''μ''. Under a simple model 4''N'' and 2''t'' are not expected to change among loci. However, the mutation rate, ''μ'', can easily change among different genes but the effect on both diversity and difference is expected to be proportional. The authors found that diversity within a species increases with rates of recombination along a chromosome, but differences between species do not. This is not consistent with the neutral theory and indicates some form of selection is acting in a general way with genome level effects. (This stands in sharp contrast to the original formulation of the neutral theory.) |
| + | |||
| + | The leading hypothesis is that diversity reducing selection is acting within species---and this affects a larger ragion around targets of selection in areas of lower recombination. The big question is what kind of selection. Both [[Hitchhiking]] from [[Selective Sweeps]] and [[Background Selection]] can contribute to this pattern and are not mutually exclusive. (This is related to the idea of [[Hill–Robertson Interference]].) Also, keep in mind that selective sweeps need not necessarily be adaptive. Often hitchhiking is framed in terms of adaptive evolution. However, "selfish" alleles can cause a hitchhiking effect without increasing an organism's fitness in a Darwinian sense. | ||
| + | |||
| + | There are additional hypotheses that can enhance the observed effect but these, in general, depend on an interaction with other forms of selection such as hitchhiking and background selection. One is that evolution favors locating genes that are more adaptive with higher levels of diversity, such as balancing selection at genes such as MHC and the ABO blood group, in regions of higher recombination, to promote maintaining diversity in the face of widespread diversity reducing selection. Another is that recombination is mutagenic (a higher mutation rate in regions of higher recombination)---there is evidence for this in some species---and selection is acting more efficiently in regions of higher recombination to remove slightly deleterious alleles before they can fix along the lineage leading to a species (there is theoretical support for this, but again it depends on an interaction with other forms of selection such as hitchhiking and background selection). These can be combined. For example, genes that undergo rapid evolution (such as those involved in immune response) might be more adaptive in regions of higher recombination because of the mutagenic effect of recombination---this can be independent of background selection and hitchhiking (but is not necessarily so)---, but this is unlikely to be generally true for many genes in the genome (most mutations, if they have a fitness effect, result in lower fitness; it is easier to break something than improve it when making random changes). | ||
[[Category:Publication]] | [[Category:Publication]] | ||
Latest revision as of 09:15, 21 October 2018
Contents
Reference
Begun, D. J., & Aquadro, C. F. (1992). Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature, 356(6369), 519.
Links
- https://www.nature.com/articles/356519a0
- http://hawaiireedlab.com/pdf/b/begunandaquadro1992.pdf (internal lab link only)
Published Abstract
TWO genomic regions with unusally low recombination rates in Drosophila melanogaster have normal levels of divergence but greatly reduced nucleotide diversity1,2, apparently resulting from the fixation of advantageous mutations and the associated hitchhiking effect3,4. Here we show that for 20 gene regions from across the genome, the amount of nucleotide diversity in natural populations of D. melanogaster is positively correlated with the regional rate of recombination. This cannot be explained by variation in mutation rates and/or functional constraint, because we observe no correlation between recombination rates and DNA sequence divergence between D. melanogaster and its sibling species, D. simulans. We suggest that the correlation may result from genetic hitch-hiking associated with the fixation of advantageous mutants. Hitch-hiking thus seems to occur over a large fraction of the Drosophila genome and may constitute a major constraint on levels of genetic variation in nature.
Notes
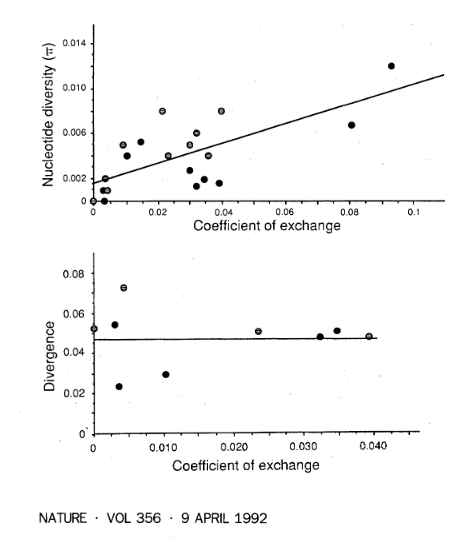
The essential result is summarized in these two figures.
Under the neutral theory diversity within a species is expected to be 4Nμ (N is the population size) and differences between species is expected to be 2tμ (t is the time of divergence in generations). Both of these are proportional to the mutation rate μ. Under a simple model 4N and 2t are not expected to change among loci. However, the mutation rate, μ, can easily change among different genes but the effect on both diversity and difference is expected to be proportional. The authors found that diversity within a species increases with rates of recombination along a chromosome, but differences between species do not. This is not consistent with the neutral theory and indicates some form of selection is acting in a general way with genome level effects. (This stands in sharp contrast to the original formulation of the neutral theory.)
The leading hypothesis is that diversity reducing selection is acting within species---and this affects a larger ragion around targets of selection in areas of lower recombination. The big question is what kind of selection. Both Hitchhiking from Selective Sweeps and Background Selection can contribute to this pattern and are not mutually exclusive. (This is related to the idea of Hill–Robertson Interference.) Also, keep in mind that selective sweeps need not necessarily be adaptive. Often hitchhiking is framed in terms of adaptive evolution. However, "selfish" alleles can cause a hitchhiking effect without increasing an organism's fitness in a Darwinian sense.
There are additional hypotheses that can enhance the observed effect but these, in general, depend on an interaction with other forms of selection such as hitchhiking and background selection. One is that evolution favors locating genes that are more adaptive with higher levels of diversity, such as balancing selection at genes such as MHC and the ABO blood group, in regions of higher recombination, to promote maintaining diversity in the face of widespread diversity reducing selection. Another is that recombination is mutagenic (a higher mutation rate in regions of higher recombination)---there is evidence for this in some species---and selection is acting more efficiently in regions of higher recombination to remove slightly deleterious alleles before they can fix along the lineage leading to a species (there is theoretical support for this, but again it depends on an interaction with other forms of selection such as hitchhiking and background selection). These can be combined. For example, genes that undergo rapid evolution (such as those involved in immune response) might be more adaptive in regions of higher recombination because of the mutagenic effect of recombination---this can be independent of background selection and hitchhiking (but is not necessarily so)---, but this is unlikely to be generally true for many genes in the genome (most mutations, if they have a fitness effect, result in lower fitness; it is easier to break something than improve it when making random changes).