Difference between revisions of "Begun and Aquadro 1992"
(→Links) |
|||
| Line 8: | Line 8: | ||
=Published Abstract= | =Published Abstract= | ||
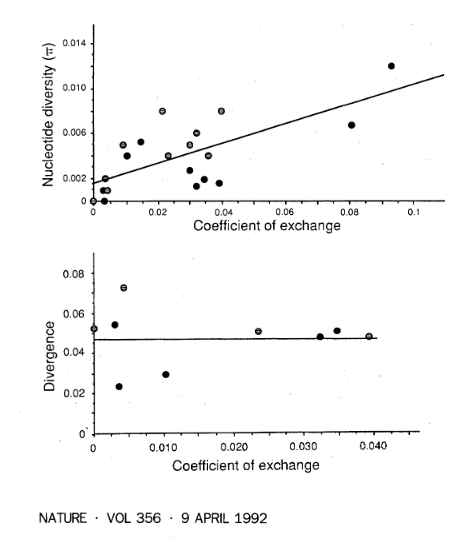
TWO genomic regions with unusally low recombination rates in Drosophila melanogaster have normal levels of divergence but greatly reduced nucleotide diversity1,2, apparently resulting from the fixation of advantageous mutations and the associated hitchhiking effect3,4. Here we show that for 20 gene regions from across the genome, the amount of nucleotide diversity in natural populations of D. melanogaster is positively correlated with the regional rate of recombination. This cannot be explained by variation in mutation rates and/or functional constraint, because we observe no correlation between recombination rates and DNA sequence divergence between D. melanogaster and its sibling species, D. simulans. We suggest that the correlation may result from genetic hitch-hiking associated with the fixation of advantageous mutants. Hitch-hiking thus seems to occur over a large fraction of the Drosophila genome and may constitute a major constraint on levels of genetic variation in nature. | TWO genomic regions with unusally low recombination rates in Drosophila melanogaster have normal levels of divergence but greatly reduced nucleotide diversity1,2, apparently resulting from the fixation of advantageous mutations and the associated hitchhiking effect3,4. Here we show that for 20 gene regions from across the genome, the amount of nucleotide diversity in natural populations of D. melanogaster is positively correlated with the regional rate of recombination. This cannot be explained by variation in mutation rates and/or functional constraint, because we observe no correlation between recombination rates and DNA sequence divergence between D. melanogaster and its sibling species, D. simulans. We suggest that the correlation may result from genetic hitch-hiking associated with the fixation of advantageous mutants. Hitch-hiking thus seems to occur over a large fraction of the Drosophila genome and may constitute a major constraint on levels of genetic variation in nature. | ||
| + | |||
| + | =Notes= | ||
| + | |||
| + | [[File:BegunAndAquadro1992Figures.png]] | ||
[[Category:Publication]] | [[Category:Publication]] | ||
Revision as of 23:25, 20 October 2018
Contents
Citation
Begun, D. J., & Aquadro, C. F. (1992). Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature, 356(6369), 519.
Links
- https://www.nature.com/articles/356519a0
- http://hawaiireedlab.com/pdf/b/begunandaquadro1992.pdf (internal lab link only)
Published Abstract
TWO genomic regions with unusally low recombination rates in Drosophila melanogaster have normal levels of divergence but greatly reduced nucleotide diversity1,2, apparently resulting from the fixation of advantageous mutations and the associated hitchhiking effect3,4. Here we show that for 20 gene regions from across the genome, the amount of nucleotide diversity in natural populations of D. melanogaster is positively correlated with the regional rate of recombination. This cannot be explained by variation in mutation rates and/or functional constraint, because we observe no correlation between recombination rates and DNA sequence divergence between D. melanogaster and its sibling species, D. simulans. We suggest that the correlation may result from genetic hitch-hiking associated with the fixation of advantageous mutants. Hitch-hiking thus seems to occur over a large fraction of the Drosophila genome and may constitute a major constraint on levels of genetic variation in nature.