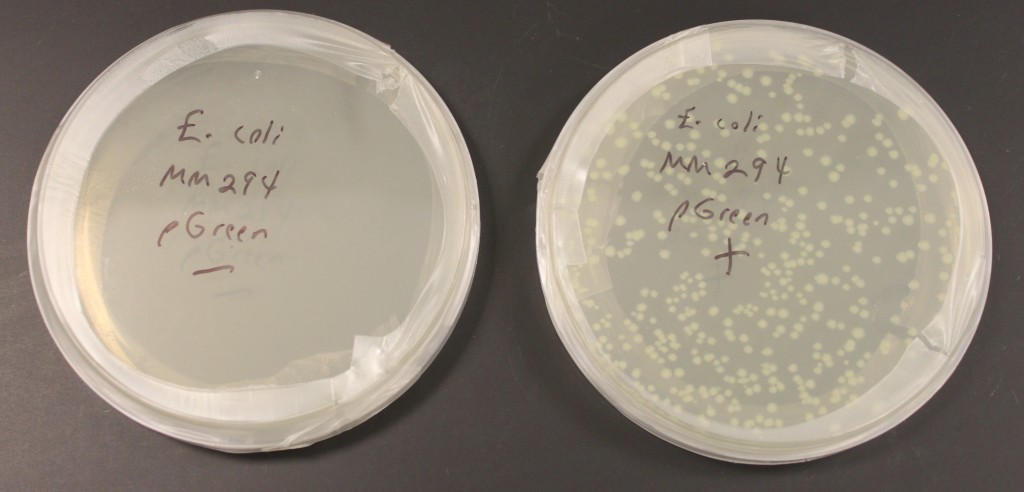
In the last few days I have been practicing transforming bacterial cells with small loops of DNA called plasmids. The picture above shows two plates of media for growing E. coli bacteria. (Escherichia coli bacteria are naturally found in our gut but some forms can also be responsible for food poisoning.) These plates also contain the antibiotic ampicillin in the media. This strain of E. coli, MM294, is not resistant to ampicillin. I mixed the bacterial cells with a plasmid containing a gene that provides ampicillin resistance in one tube, and in a control tube I did not add any of the plasmid. This was done in a calcium chloride solution (CaCl2) cooled on ice that helps the DNA associate closely with the cell membranes. The vials are then "heat shocked" in 42 C water, which disrupts the cell membranes and helps the plasmid to be taken into the cell. Then I spread the cells on the plates to grow overnight. In the left "-" plate without the plasmid added the ampicillin killed the cells. In the right "+" plate the "amp" resistance gene on the plasmid allowed cells to grow and divide that had taken up the plasmid. Each spot is the descendants of a single original cell that was transformed (within a spot the cells are essentially genetically identical and are referred to as "clones"). The plasmid also contains a modified green fluorescent protein (GFP) from a bioluminescent jellyfish (Aequora victoria) that causes them to faintly glow green under UV light. The GFP modification, a fusion of part of another gene, also gives them a light green color in regular light that you can see above. (pGreen refers to this plasmid.)
Bacteria Transformation, pGreen
2 Replies