First of all there has been a long lag in posting any updates here about the lab. There are many things going on that are a higher priority at the moment; however, I also want to try to keep a certain tempo of posts and updates going here if possible. So, I set aside a few minutes today to make a post.
A couple of undergraduates in the lab have an interest in viruses, specifically bacteriophages. They have worked out methods to isolate and propagate the phages for various experiments. However, we wanted to get a look at the actual viral particle instead of inferring it indirectly from plaques on a plate of bacteria. I talked to Tina Carvalho at our Biological Electron Microscope Facility and she was enthusiastic about imaging the viruses.
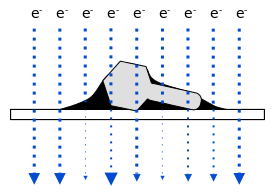
Negative staining is a relatively quick and easy method to get viral mugshots. The virus particles are put onto a thin membrane held in a metal grid and electrons are beamed through both the virus and the grid to get an image. The problem is that this doesn't really work that well. The virus is very transparent to electrons and they will pass right through almost like it wasn't there, i.e., the image contrast defining the virus will be low and look faint.
A stronger image can be made by adding a thin fluid of uranium salt (Uranyl acetate (UO2(CH3COO)2·2H2O)). Uranium is very dense and good at blocking electrons. If all goes well (and Tina pointed out that there can be quite a bit of luck involved with the color of you socks that day possibly playing a role) the fluid will dry a bit and form droplets around the virus particles by adhesion. The electrons pass through the virus, which is making a path of sorts, through the uranium drop, so the outside shell of the viral surface can be better resolved. Here is a figure to try to illustrate.
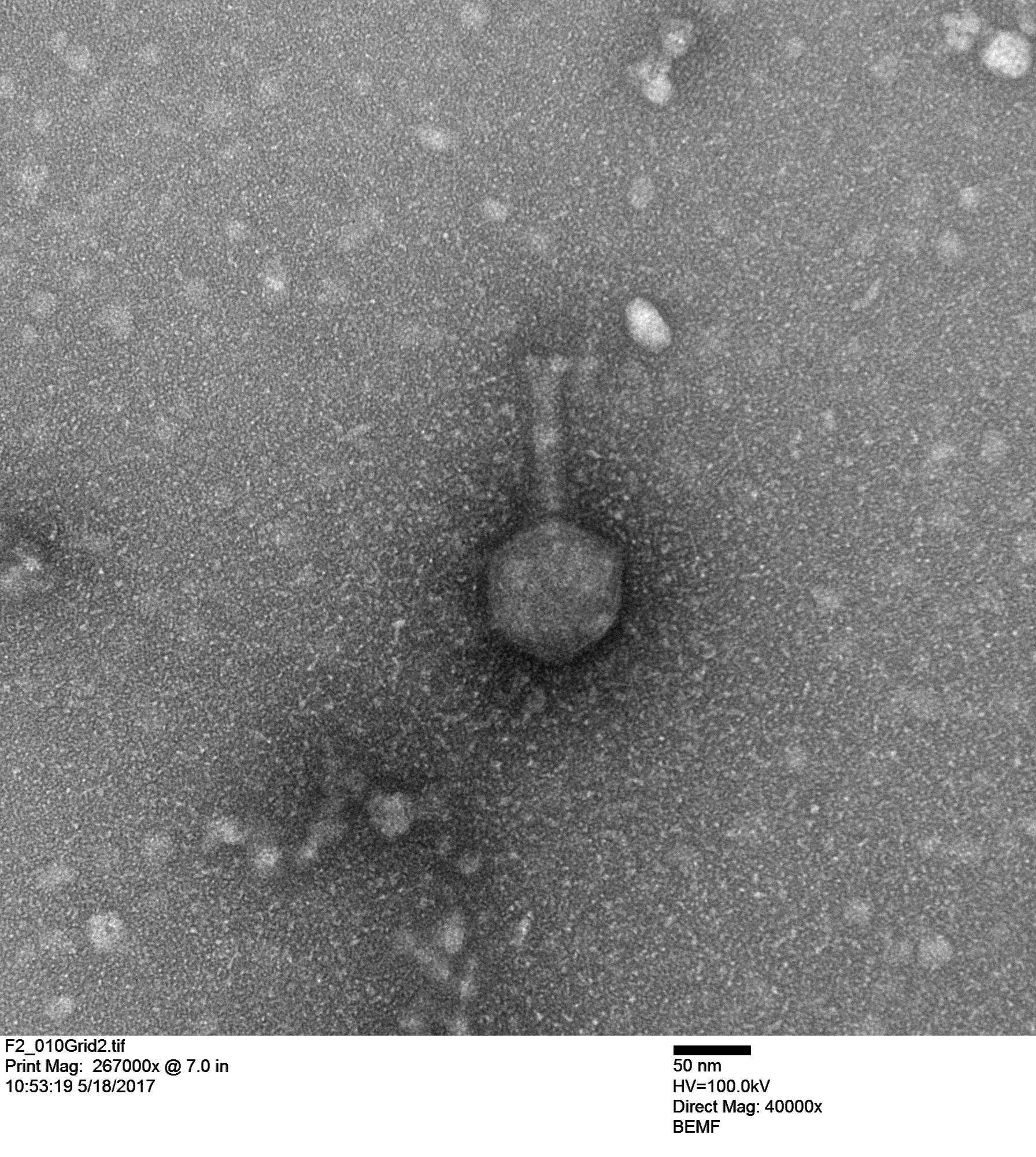
First we looked at a Vibriophage that Stacy Paulino isolated from seawater from Maunalua Bay. This phage infects a coral pathogen, Vibrio coralliilyticus, and Stacy imaged it below.
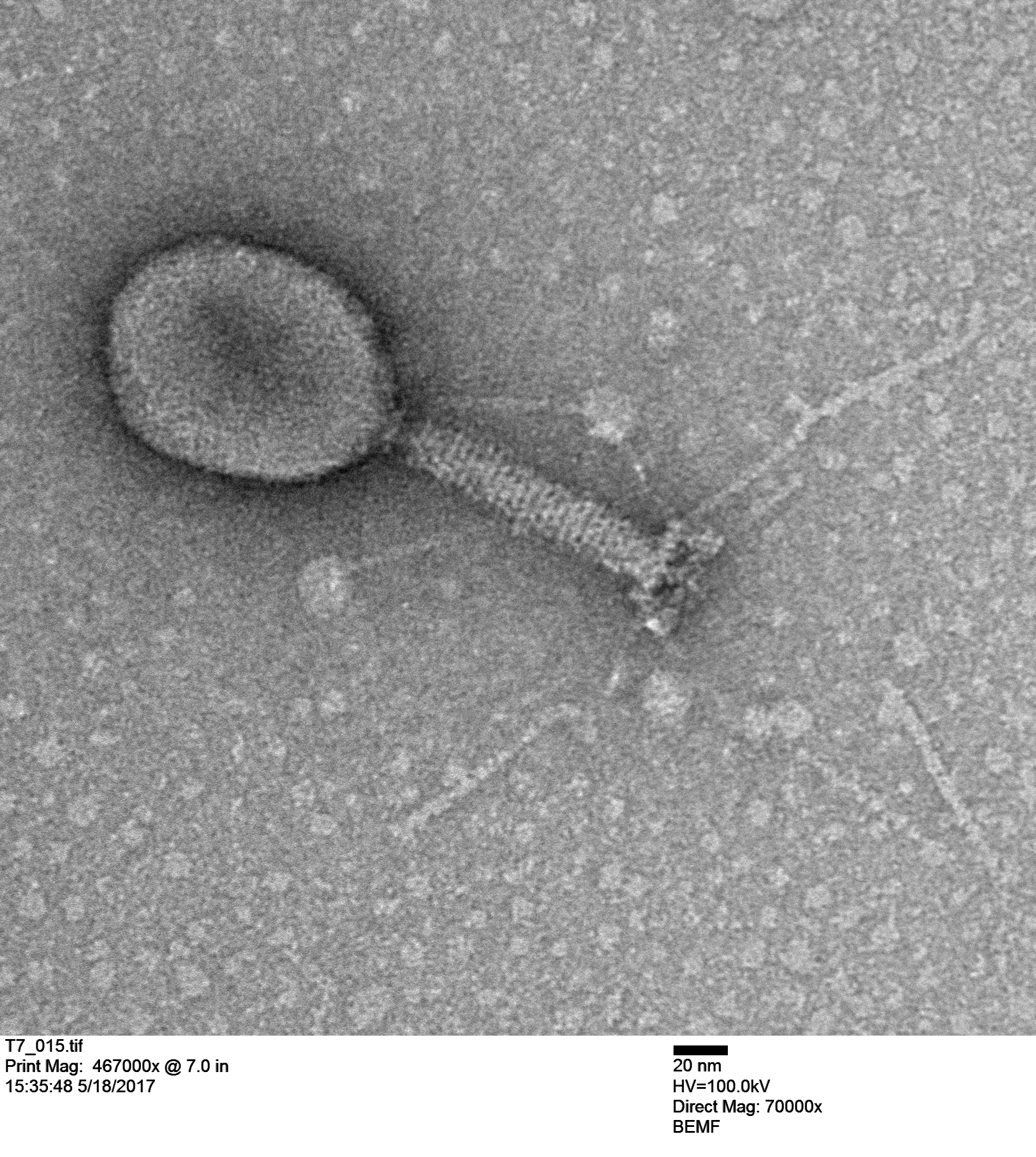
The next image is an unknown coliphage (infects Escherichia coli).
Notice the scale in the image indicating 20 nanometers; that is the size of a ribosome. Double stranded DNA is about 2 nm wide of 1/10 of the scale bar. The graininess of the tube is from individual tail tube proteins, single polypeptide molecules, which are typically assembled from ~250 individual peptides.
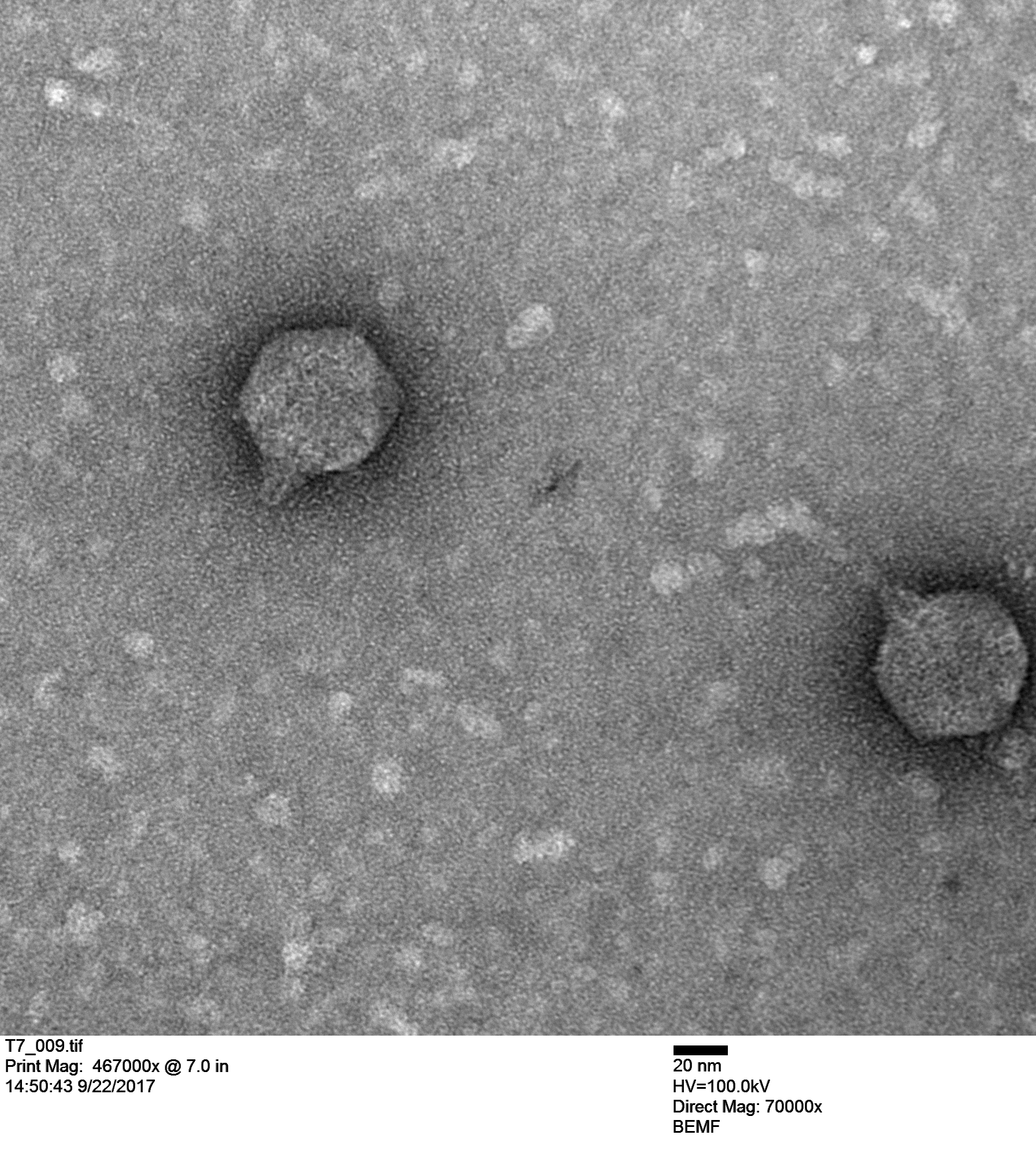
Maya Shaulsky (in a collaboration with Bob Thomson) is working with T7 (below) and made these coliphage images. Originally we thought we were working with T7 coliphage but we kept getting frustrating results that didn't make sense. When we actually saw the virus (the image above) it confirmed that we were not working with T7 so we ordered actual T7 from a different stock center.
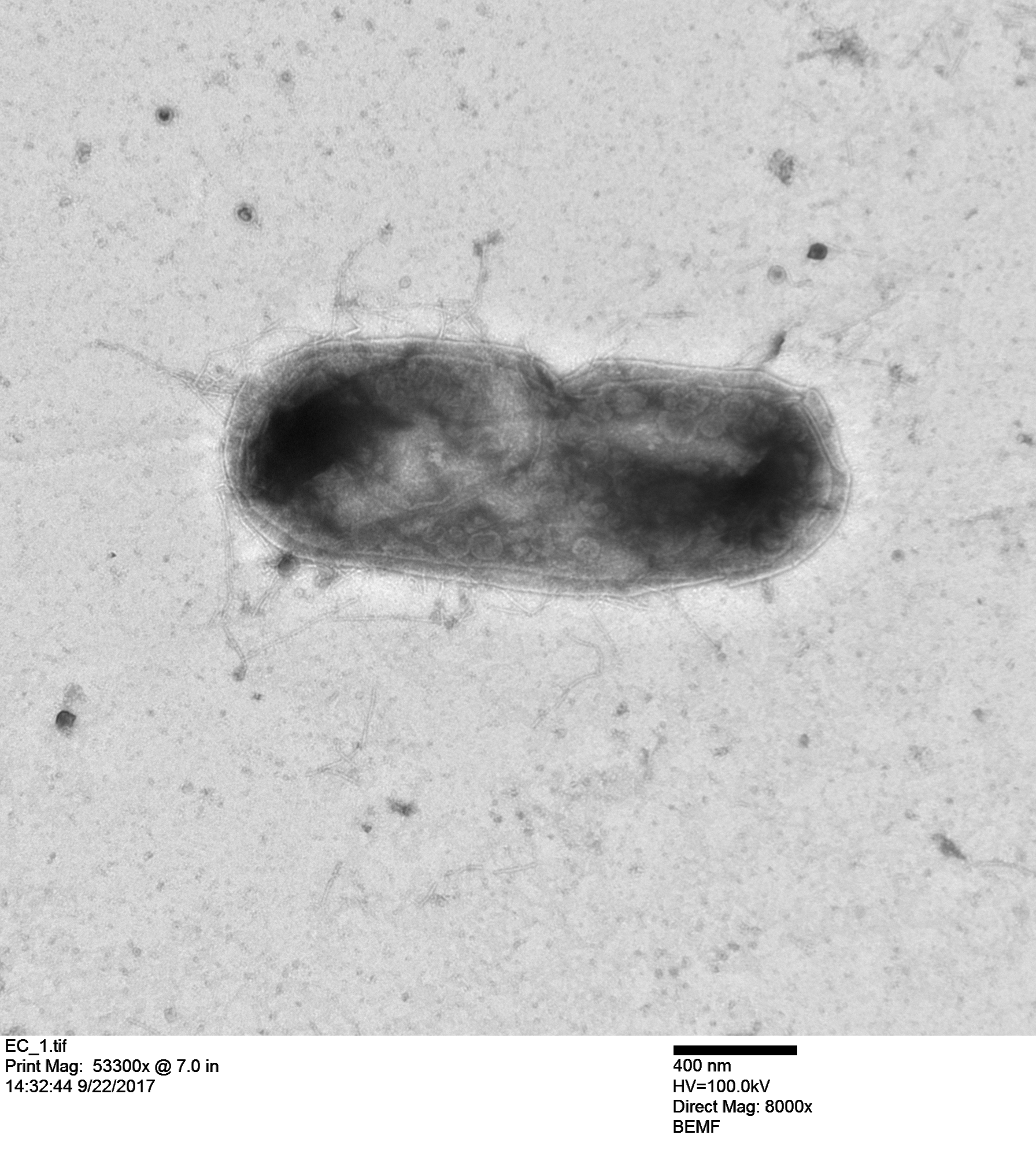
And here is an E. coli bacterium with T7 particles replicating inside the cell!
If you are working with something small I encourage you not to be intimidated with the idea of electron microscopy imaging. It is really something to actually see what you are working with. The protocol to prepare the specimens is not that long or difficult (some people may disagree with this, perhaps we have been lucky) and it is not really that expensive to pay for the training and machine time. It is worth trying at least once.