The last post about my COI "species barcode" sequence has been bugging me. I wouldn't really expect to find a unique sequence in a small region by chance in a mitochondrial gene. Many, many mitochondria in humans have been sequenced and the general levels of genetic variation are well understood. I checked closer and it appeared to be an amino acid altering mutation to a different class of amino acid--not expected at all.
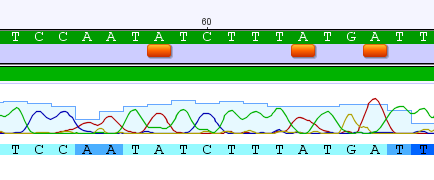
It was close to one edge of the sequence so, I looked at the other edge and found some more unique changes. In the sequence snippet above each A that is underlined by an orange box is a G in standard human COI sequences. I made an assembly with the human reference sequence and added my primers to the alignment. Then it became obvious that I had made a rookie mistake. The "mutations" were located in the primer. These primers were not designed to only work with humans, but to work across a wide range of animals. They do not match the human sequence exactly. As the PCR progressed making more and more copies starting from the annealed primers the primer sequence was incorporated into the total sequence.
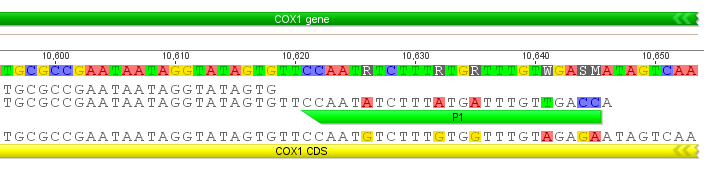
The image above shows the reference sequence at the bottom, the primer position is in light green and my sequence is above that. The changes match the primer (5'-GGTCAACAAATCATAAAGATATTGG-3' the compliment of which is 5'-CCAATATCTTTATGATTTGTTGACC-3' in the sequence above). This is actually a method to engineer specific changes to a DNA sequence, a form of site directed mutagenesis.
Going back to the first side and taking another look:
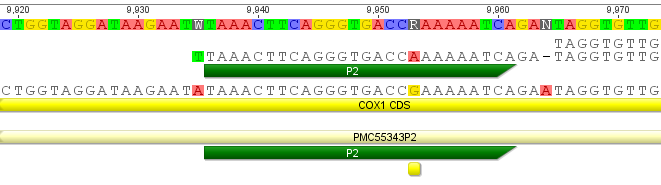
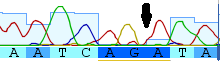
The A is incorporated from the primer (5'-TAAACTTCAGGGTGACCAAAAAATCA-3'). There is also a missing A just inside the sequence (in the 3' direction); taking a closer look at the trace I can agree that an extra A may be at the position (the arrow below points to a "shoulder" on the red A trace that may be the missing nucleotide):
There is also a "T" just outside (5' to) the primer sequence.
The Taq polymerase enzyme used in PCR is known to be error prone and sometimes add A's on to the 3' end of a PCR product (in fact this is exploited in one method, TA cloning, to clone PCR products). By convention sequences are written 5' to 3' so a 3' A will appear as a complementary T in this instance. also, note in the previous figure with the first primer an extra A was present on the 3' end (but this also agrees with the reference sequence).
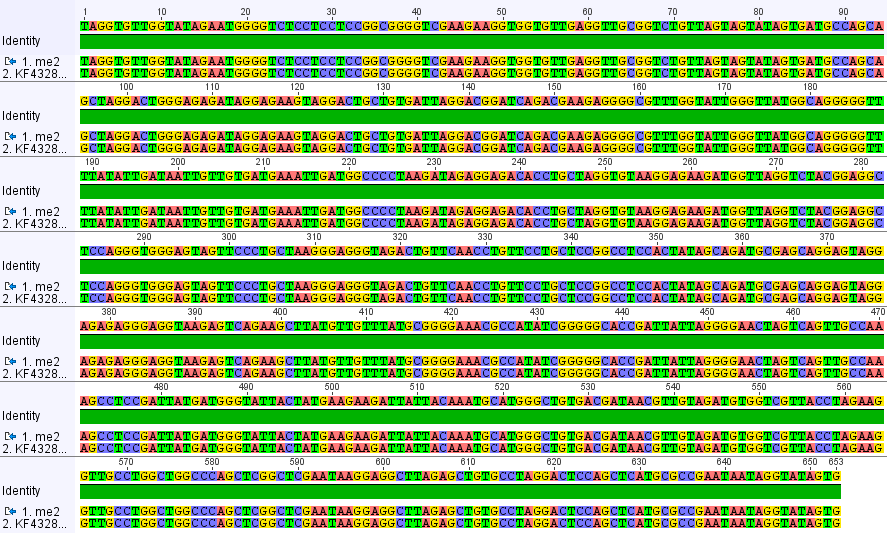
When I trimmed out the edges and carefully curated the sequence I got the following alignment below.
100% identical to many human sequences on genbank.
So, just for the record, my corrected COI DNA sequence, in FASTA format is below:
>me_COI TAGGTGTTGGTATAGAATGGGGTCTCCTCCTCCGGCGGGGTCGAAGAAGGTGGTGTTGAGG TTGCGGTCTGTTAGTAGTATAGTGATGCCAGCAGCTAGGACTGGGAGAGATAGGAGAAGTA GGACTGCTGTGATTAGGACGGATCAGACGAAGAGGGGCGTTTGGTATTGGGTTATGGCAGG GGGTTTTATATTGATAATTGTTGTGATGAAATTGATGGCCCCTAAGATAGAGGAGACACCT GCTAGGTGTAAGGAGAAGATGGTTAGGTCTACGGAGGCTCCAGGGTGGGAGTAGTTCCCTG CTAAGGGAGGGTAGACTGTTCAACCTGTTCCTGCTCCGGCCTCCACTATAGCAGATGCGAG CAGGAGTAGGAGAGAGGGAGGTAAGAGTCAGAAGCTTATGTTGTTTATGCGGGGAAACGCC ATATCGGGGGCACCGATTATTAGGGGAACTAGTCAGTTGCCAAAGCCTCCGATTATGATGG GTATTACTATGAAGAAGATTATTACAAATGCATGGGCTGTGACGATAACGTTGTAGATGTG GTCGTTACCTAGAAGGTTGCCTGGCTGGCCCAGCTCGGCTCGAATAAGGAGGCTTAGAGCT GTGCCTAGGACTCCAGCTCATGCGCCGAATAATAGGTATAGTG
Incidentally, this section is now an exact match to the Cambridge reference sequence, NC_012920.