My recent news about being promoted with tenure has emboldened me to write about some things that I have kept pent up for a long time. I'm not sure if this is a good thing but lets see where this leads us.
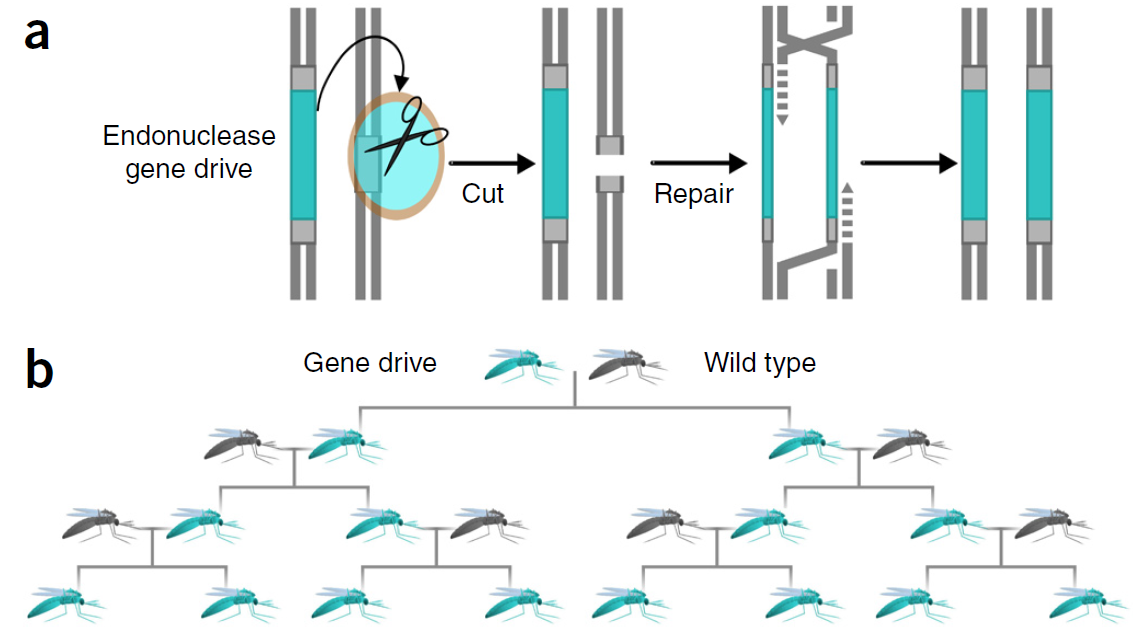
We engineered and demonstrated a self-limiting gene-drive for local and reversible genetic modification of a population. There is an argument about whether this type of system, underdominance, should even be considered gene-drive in the broader sense---but I am not going to go into definition arguments here. It is certainly a much safer type of population transformation system than alternatives that can invade a population from arbitrarily low frequencies.
Dr. R. Guy Reeves and I first began discussing engineered haploinsufficient mediated underdominance in 2006 when I returned from some work in Africa. I hired him as a postdoc in my new position at the Max Planck Institute for Evolutionary Biology in 2008 and we began work engineering the system. We knew it would take years to troubleshoot the technology so we also published theoretical results to lay the groundwork for making predictions of underdominant systems, e.g. Altrock, P. M., Traulsen, A., Reeves, R. G., & Reed, F. A. (2010). Using underdominance to bi-stably transform local populations. Journal of Theoretical Biology, 267(1), 62–75. doi:10.1016/j.jtbi.2010.08.004
We made three inserts into the genome of Drosophila melanogaster of our engineered genetic construct and started testing them. How do you test if you have generated underdominace? This requires tracking the frequency of the insert over multiple generations in multiple replicate populations, which takes time and is quite a bit of work. I spent many nights in the lab counting thousands of flies and then walking home for a couple hours of sleep before sunrise. The first insert was homozygous lethal and useless for engineering underdominance. The second insert had lower homozygous fitness than as a heterozygote (technically a hemizygote) and did not result in underdominance. The third insert was interesting. As I collected data and each generation went by it began to look more and more like underdominance and became more and more statistically significant. I remember late one day in the winter of 2009/2010 Guy was going to leave to take the train back home to Hamburg and I asked him to stay and catch the next train. I had a notebook full of new data and I wanted him to see how it came out when I plotted it and did the calculations. It was clear unambiguous underdominance! I presented the results at our next meeting in May 2010 (Reed, F. A. and R. G. Reeves. Underdominance theory meeting data, how do they get along? Aquavit VIII meeting, The Max Planck Institute for Evolutionary Biology, Plön, Germany. slides PDF link). I also presented the results at some other talks such as February 2011 in Hawai'i (slides PDF link). Obviously we were excited about this, wrote up our results, and submitted them for publication!
All along we were aware of the potential for unintended dynamics of the system; I mentioned just this concern in a publication in 2007 (Reed, F. A. (2007). Two-locus epistasis with sexually antagonistic selection: A genetic Parrondo’s paradox. Genetics, 176(3), 1923–1929. doi:10.1534/genetics.106.069997). We were aware of invasive Medea gene-drive dynamics and discussed the possibility of (unintended) maternal deposition of the RNA "poison" into embryos with rescue depending on transcription in the embryos---this could completely change the dynamics of the system. So, we built a fail-safe into our system. We divided expression control of the RNA "poison" over two chromosomes, that have to be together in the same individual for it to work, using a standard binary control system (GAL4/UAS). If genetically modified "gene-drive" flies escaped from the lab then independent assortment (male recombination is suppressed) of the chromosomes in the following generation would break the system and it would not be able to drive. This enabled safe testing in the lab and the binary control system was not required for actual future applications of the technology where the fail-safe could be removed from the system (we went to great pains to explain this to reviewers, backed up with facts about how our flies were transformed).
So, it turns out that we were unknowingly in competition with another lab to be the first to publish a self-limiting gene drive system. When we submitted our manuscript for publication we encountered a very hostile reviewer. (There were also other issues at play that delayed the process, the Max Planck system decided to pursue a patent on the technology, there were personalities involved, etc. However, the reviews were truly maddening and am what I am focusing on here.) This person tried to find a reason to reject the manuscript and focused on the fail-safe---claiming that our approach could not work without this in place. We were eventually rejected from publishing in journal after journal, and the hostile reviews followed us from journal to journal, in some cases with the exact same review copied from the previous journal submission despite our revisions to the manuscript. This dragged out over a period of years and then we were finally able to publish in PLoS ONE (Guy Reeves, R., Bryk, J., Altrock, P. M., Denton, J. A., & Reed, F. A. (2014). First steps towards underdominant genetic transformation of insect populations. PLoS ONE, 9(5). doi:10.1371/journal.pone.0097557). However, Akbari et al. was published in 2013 (Akbari, O. S., Matzen, K. D., Marshall, J. M., Huang, H., Ward, C. M., & Hay, B. A. (2013). A Synthetic Gene Drive System for Local , Reversible Modification and Suppression of Insect Populations. Current Biology, 23(8), 671–677. doi:10.1016/j.cub.2013.02.059). Even though their names appear here, this is not an attack directed at Akbari, Hay, or anyone else named here; I do not know the names of the reviewers of our manuscript (and later our grant applications) and I am not implying that it is any of these people in particular.
Our system has a lot of potential advantages, not least of which is the likely species portability of the approach due to the ubiquity of haploinsufficiency of ribosomal proteins across species. The Akbari et al. 2013 approach depends on careful control of expression timing during development, and while it certainly could be ported across species this is likely to be more difficult. However, we seem to have been blackballed. When applying for grant funding to implement this system in mosquitoes I get comments back the reflect some of the hostile reviews we received earlier (that this system is "fanciful" and cannot work in the wild, etc.). I have seen presentations where Akbari et al. 2013 is credited with the first self-limiting gene drive (no, we presented our results in 2010, 2011, we have a patent priority date of 2012). And I see reviews where our system is described as only proof-of-principle while the Akbari et al. 2013 system is described in contrast as a "fully functional system capable of invading wild populations" (Champer, J., Buchman, A., & Akbari, O. S. (2016). Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nature Reviews Genetics, 17(3), 146–159. doi:10.1038/nrg.2015.34) ... wild populations of what, Drosophila melanogaster? Transforming D. melanogaster is not useful, being able to transform other species, such as mosquitoes, is what is useful. Furthermore, accidentally transforming the entire Drosophila melanogaster species is dangerous, for reasons that not least of which it is a useful model organism, a human commensal, and because of the potential public backlash this could cause.
In another publication that was also delayed for years by a hostile reviewer, perhaps even the same person, we recommended combining underdomiannce with gene-drive systems like Medea in order to protect laboratory model organisms from unintended species-wide genetic modifications (Gokhale et al. 2014, http://bmcevolbiol.biomedcentral.com/articles/10.1186/1471-2148-14-98). The point I am trying to make is that being thoughtful and safely designing gene-drive systems, with safety checks and fail-safes in place, should be encouraged rather than discouraged within the scientific community. Unfortunately, in my experience the opposite seems to be true.