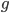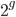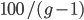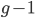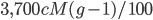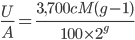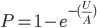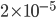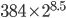I found a link with a relative that has me a bit surprised. I share two segments on two different chromosomes with "J". We compared genealogies and we have common ancestors, but they are further back than I expected. Nine generations back J's 6X-great-grandfather John Gillett (1644-1682) is a brother to my 7X-great grandmother Mary Gillett (1637-1719). J and I are linked via a path of 19 generations; this is a family in the Connecticut colony in the 1600's!
One caveat to add here is that both sides married into the Barber family of Connecticut (with an unknown but perhaps likely connection between the Barbers) in the next generation, so the connection may be slightly closer (in a genetic sense) than it first appears. For the sake of argument lets consider this a path of 18 generations back and forth through time; I am still surprised at the time depth.
It is possible that we are connected via another closer unknown common ancestor, but we appear to both have well worked out genealogies and after Connecticut there is not any apparent overlap in locations the families moved through or surnames that are shared.
This got me to thinking about just what kind of connections we do expect over the last 10 generations or so...
Ignoring inbreeding in the genealogy sense, our number of ancestors doubles each generation we go back (2 parents, 4 grandparents, 8 great-grandparents, 16, 32, 64, 128, 256, 512, 1024 (ancestors 10 generations back), ...). (With inbreeding it levels off to something like the effective population size after a sufficient number of generations.) The number of our genealogical ancestors grows exponentially back in time, but our genome is finite in size, so something has to give. The result is that some ancestors start to drop out from the genetic representation that we have inherited in our genome.
So, to write this down, if 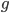 is generations we have
is generations we have 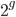 ancestors each generation back. A Morgan (M) is a unit of genetic recombination. In one generation we expect, on average, one recombination event per Morgan distance along the chromosome. Usually this is reported in units of centi-Morgans (cM), where one Morgan is equal to 100 cM. This suggests that our genome is whittled up into units of
ancestors each generation back. A Morgan (M) is a unit of genetic recombination. In one generation we expect, on average, one recombination event per Morgan distance along the chromosome. Usually this is reported in units of centi-Morgans (cM), where one Morgan is equal to 100 cM. This suggests that our genome is whittled up into units of 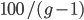 cM. The
cM. The 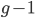 term is there because we inherit an entire genome copy from each parent and recombination affects the generation before--our grandparents.
term is there because we inherit an entire genome copy from each parent and recombination affects the generation before--our grandparents.
So, if our entire genome is approximately 3,700 cM long (ignoring the breaks between chromosomes and to simplify, pretending for the moment that the entire genome is linked on one continuous chromosome). Then we expect 37 units of 100 cM length (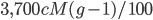 from our grandparents, 74 units from our great grandparents, etc.
from our grandparents, 74 units from our great grandparents, etc.
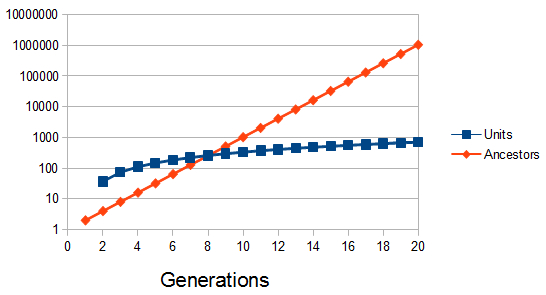
We can divide this by the expected number of ancestors each generation to get an average number of genetic units per ancestor: 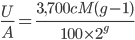
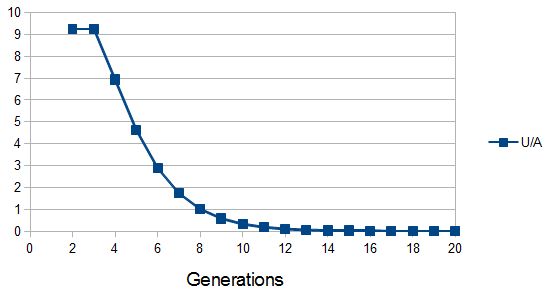
Of course there is a lot of variation. Each unit is not of the same size and number from each ancestor, this is just an average expectation. If we consider ancestral representation as a Poisson process with the expectation as a mean of the Poisson distribution we can plot the probability that an ancestor is represented at least once in our genome, which is one minus the probability they are represented zero times. 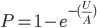
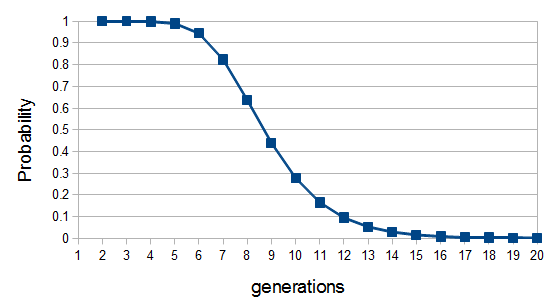
So up to five generations back we inherit parts of our genome from ancestors with near certainty, then there is a steep drop off between six to twelve generations. The chance that we have inherited anything from a specific ancestor twenty generations back (or 600 years ago assuming an average of 30 years per generation) is practically zero (but of course we did inherit each segment from someone twenty generations back). Most of our family connections should be within a connection of about 14 generations or a common ancestor around seven generations (our great-grandparents, grandparents, grandparents; ~200 years) back.
So what about the probability of a common identical-by-descent (IBD) chromosomal segment preserved over 18 generations (or from a common ancestor ~9 generations ago)? Actually, since they are full sibs (sharing both parents) rather than half sibs we should subtract another generation to reflect that there are two ways to be genetically related--through the mother or father. According to the calculations above the chance of inheriting a segment from an ancestor eight or nine generations ago is 64% and 44% respectively. The chance of both of us sharing the track from the common ancestor, over an adjusted 17 generations, is 0.45% or about one out of 220 with an expected length of 6.25 cM. We actually share two tracks on two different chromosomes, which makes the combined probability something like 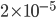 or one out of 50,000.
or one out of 50,000.
So this match seems to be very unlikely. However, there is a important issue to bring up here. I did not pick a specific relative, descended from a specific common ancestor, and compare our genomes to see if we had any matches. If that were the case then the calculation above is appropriate and this is very unexpected. What actually happened was that any of the many tens to hundreds of thousands of relatives who might have had a match between our hundreds of common ancestors eight to nine generations ago were picked and the ones I did not have a match with were not. (The average of 215 and 512 ancestors eight to nine generations ago is 384. Assuming a family of four children per ancestor pair, two per person, gives 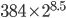 or about 140,000 relatives.) So, if something has a probability of one out of 50,000 but we have over 140,000 chances then we actually expect it to occur.
or about 140,000 relatives.) So, if something has a probability of one out of 50,000 but we have over 140,000 chances then we actually expect it to occur.
However, there is a counteracting force at work here as well. In fact the main limitation is likely the number of people in 23andme's database to compare to. It is over 100,000 which corresponds to more than one per every 3,000 people in the US (for simplicity assume the vast majority of 23andme customers are in the US). So, only about 0.03% of my relatives have been sampled, which reduces the rough estimate of 140,000 chances in the preceding paragraph to an effective 46 chances. This makes the one out of 50,000 odds something like one out of 1,000, which is not expected.
So, I would not be surprised if we later find another common ancestor that we had missed before.
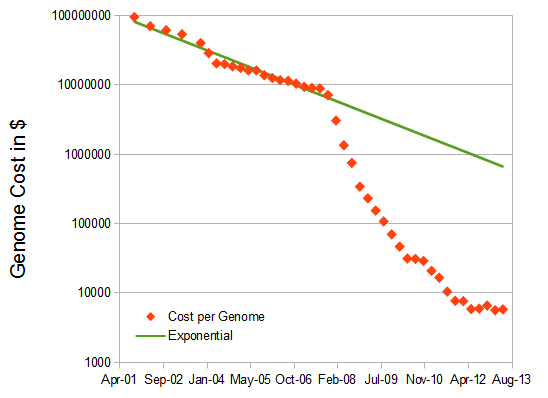
 . However, after 2007 new, massively parallel, sequencing technologies came on the scene (454, Illumina, SOLiD) that drove the price down at an unprecedented rate. Now there is a rush to the $1,000 genome.
. However, after 2007 new, massively parallel, sequencing technologies came on the scene (454, Illumina, SOLiD) that drove the price down at an unprecedented rate. Now there is a rush to the $1,000 genome.