Unusual heterokaryon complementation patterns
When heterokaryons are produced the cells fuse but the nuclei do not. This might provide an opportunity to study effects that are restricted to the nuclei versus effects that are shared between nuclei. mRNA is exported out of the nucleus to the rough endoplasmic reticulum and protein products are imported back into the nucleus through the nuclear pore complex if they have the correct nuclear localization signal. Proteins can be quickly shared among the nuclei of heterokaryons (e.g., Piñol-Roma and Dreyfuss 1992, also see the discussion of Rao et al. 1970 (who worked out the cell cycle process from signals shared among heterokayon nuceli) here http://www.nature.com/celldivision/milestones/full/milestone06.html).
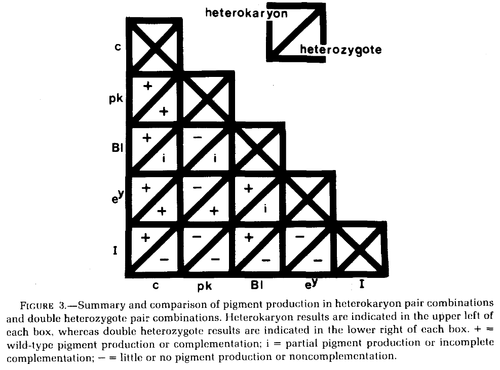
Wilkins et al. 1982 studied variants at five genes affecting chicken pigmentation (recessive white c, pink-eye pk, blue Bl, recessive wheaten ey, and dominant white I) in both double heterozygotes and in heterokaryons.
In the chart above Bl is incompletely dominant in heterozygotes and therefore does not fully restore a wildtype phenotype.
Dominant white I does not complement in double heterozygotes (because of dominance) but does complement with c and Bl in heterokaryons, perhaps suggesting its function is restricted to the nucleus. However, dominant white I is a 9bp insertion mutation in exon 10 of PMEL17, a transmembrane protein required for eumelanosome development (Kerje et al. 2004).
Similarly, pk does not complement with Bl or ey, again perhaps suggesting function limited to the nucleus. pk may be almost lost in chicken stocks (http://stalderexotics.com/chicken_genetics.htm) but may be "P protein" (corresponding to the gene OCA2) which is a melanosome anion pump (Brilliant 2001; http://www.uniprot.org/uniprot/Q04671).
Two mechanisms come to mind to help explain how two nuclei that share a cytoplasm would have functions restricted to within the nucleus. One is possible nuclear architecture interactions that are disrupted back to a wild-type phenotype in heterozygotes; however this does not appear to be the case in the I and pk examples. Another more likely explination is perhaps regulatory RNAs. There are many RNAs that do not code for a protein but aid in regulating the expression of other genes. If the microRNA is not exported out of the nucleus it may not affect the other nucleus in a heterokaryon. microRNAs bind to mRNAs and reduce the corresponding gene expression. Perhaps the 9bp insertion in PMEL17 disrupts RNA binding and results in over-expression, which may explain dominant white I. Perhaps a similar RNA based form of regulation also explains non-comlementation in pk.
Followup reading: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4377836/ http://jcs.biologists.org/content/joces/113/1/11.full.pdf https://www.ncbi.nlm.nih.gov/pubmed/15965474