Cohen-Boyer-Berg experiment
In 1973 the first living genetically engineered transgenic organism combining DNA across taxonomic domains (bacteria and eukaryota) was made in the Cohen-Boyer-Berg experiment, which built upon a rapid series of experiments of manipulating and introducing DNA into bacterial cells.[1][2][3][4]
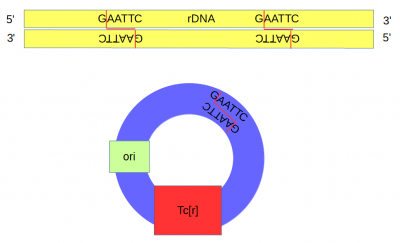
A plasmid (pSC101[5]) containing a gene conferring resistance to tetracycline (Tcr) and an origin of replication was cut at a single site with the restriction enzyme EcoRI.
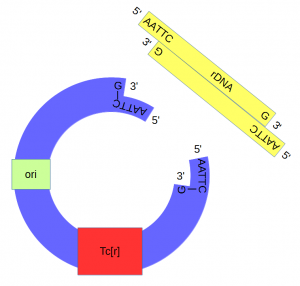
DNA from an African clawed frog (Xenopus laevis) was also cut with EcoRI and this yielded a range of DNA fragments including a large number of ribosomal DNA (that codes for rRNA) fragments[6] with matching "sticky ends".
Above, the sites that are recognized and cut by EcoRI are shown by DNA base letters and red lines respectively. Below is a schematic of the cut DNA showing the overhanging "sticky ends".
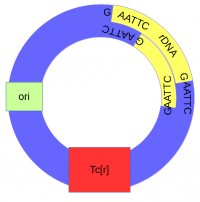
The cut plasmid and Xenopus DNA were mixed together with a ligase to repair the cut site, incorporating the Xenopus rRNA fragment into a fraction of the plasmids.
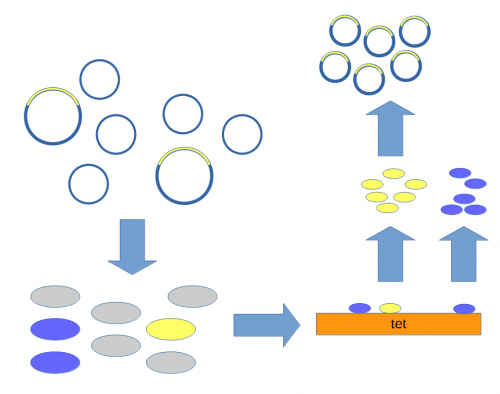
A population of E. coli cells were then transformed to incorporate the plasmids into their cells and grown on media containing tetracycline to select for the cells that contained the plasmid conferring tetracycline resistance. These could then be screened to identify the cells with plasmids that had incorporated the Xenopus DNA.
Above, the mixture of plasmids with and without the insert are introduced into E. coli cells (using calcium chloride heat shock transformation[7]). The cells are then raised on media containing tetracycline so that cells without a plasmid (gray) do not grow.[8] Individual colonies can be picked and tested for the presence of the insert (yellow) or just the plasmid without the insert (blue). Once a colony is found that contains the insert it can be grown in large numbers[9] and the DNA purified to yield the inserted plasmid.
The end result was a bacteria that stably replicated a fragment of amphibian ribosomal DNA.
References and Footnotes
- ↑ Cohen, S. N., Chang, A. C., Boyer, H. W., & Helling, R. B. (1973). Construction of biologically functional bacterial plasmids in vitro. Proceedings of the National Academy of Sciences, 70(11), 3240-3244.[1]
- ↑ Chang, A. C., & Cohen, S. N. (1974). Genome construction between bacterial species in vitro: replication and expression of Staphylococcus plasmid genes in Escherichia coli. Proceedings of the National Academy of Sciences, 71(4), 1030-1034.[2]
- ↑ Morrow, J. F., Cohen, S. N., Chang, A. C., Boyer, H. W., Goodman, H. M., & Helling, R. B. (1974). Replication and Transcription of Eukaryotic DNA in Esherichia coli. Proceedings of the National Academy of Sciences, 71(5), 1743-1747.[3]
- ↑ Cohen, S. N. (2013). DNA cloning: A personal view after 40 years. Proceedings of the National Academy of Sciences, 110(39), 15521-15529.[4]
- ↑ The plasmid is named pSC101 because it was the 101st plasmid isolated by Stanley Cohen.
- ↑ Ribosomal DNA sequences are present in many copies in the genome and in Xenopus these contain two EcoRI sites close together.
- ↑ Mandel, M., & Higa, A. (1970). Calcium-dependent bacteriophage DNA infection. Journal of molecular biology, 53(1), 159-162. [5]
- ↑ Cohen, S. N., Chang, A. C., & Hsu, L. (1972). Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proceedings of the National Academy of Sciences, 69(8), 2110-2114. [6]
- ↑ The cells are theoretically genetically identical to each other so this is referred to as "cloning" the DNA of interest.