 When looking at a map of biochemical pathways (a small part of which is above) one can start to get an idea of how complex a cell is. The protein products of genes carry out these steps to keep the chemistry of a living cell going. Disrupting these pathways by blocking a step with a gene mutations often results in a phenotype and/or in humans what we would recognize as a genetic disease. The genes are turned on and off by the expression of other genes that respond to each other and to biochemical and physical signals in the cell and from the environment by a complex regulatory logic. Furthermore, certain phenotypes and genetic diseases are also caused by, not blocking a biochemical step, but by carrying it out in the wrong place or time during development or in response to environmental stress, etc.
When looking at a map of biochemical pathways (a small part of which is above) one can start to get an idea of how complex a cell is. The protein products of genes carry out these steps to keep the chemistry of a living cell going. Disrupting these pathways by blocking a step with a gene mutations often results in a phenotype and/or in humans what we would recognize as a genetic disease. The genes are turned on and off by the expression of other genes that respond to each other and to biochemical and physical signals in the cell and from the environment by a complex regulatory logic. Furthermore, certain phenotypes and genetic diseases are also caused by, not blocking a biochemical step, but by carrying it out in the wrong place or time during development or in response to environmental stress, etc.
When thinking about this it is easy to believe that cells are highly evolved (which they are) and that making random changes to the system would almost universally result in negative effects (which ... strangely may not be as true as we might think). I like to use a car as an model of a cell in class in various ways. If you know what you are doing you can repair a car to restore its function or even add new functions. However, if you make random changes to a car, even if you just limited yourself to a single system like rewiring the electrical system or shuffling mechanical parts around in the drive train, you are very likely to, if there is any effect at all, mess the car up and render it useless (very rarely you might accidentally improve things). Using this analogy it is intuitive that shuffling the part of the gene that codes for an RNA or protein product around with the part of the gene that controls its expression (the promoter in a broad sense including regulatory regions that increase or decrease expression by interactions with other molecules) would render the cell useless, in other words result in severe phenotypes and/or lethality.
This perspective is why this article is so interesting to me,
Isalan, M., Lemerle, C., Michalodimitrakis, K., Horn, C., Beltrao, P., Raineri, E., … Serrano, L. (2008). Evolvability and hierarchy in rewired bacterial gene networks. Nature, 452(April), 840–845. doi:10.1038/nature06847.
These authors focused on transcription factors, which are sort of master switches in the cell, the gene products of transcription factors turn other groups of genes on and off. They reshuffled 26 promoter regions with 23 regulatory genes in E. coli (to put this in perspective only nine transcription factors control half of all of the genes in E. coli) and tried 598 possible combinations in a high copy plasmid (small extra chromosome present in many numbers) that was cloned (added to the cell).
In the car analogy parts were not removed and replaced with alterations; rather altered parts were added. Like adding a fifth wheel in a random orientation somewhere along the drive chain, or adding extra wires connected to a random location to the electrical system---still not a good idea for a car. In addition, within the cell some of these new combinations are predicted, based on simplistic understanding of the cellular network, to result in run-away positive or negative feedback loops when interacting with the cells normal machinery (e.g., expression of a gene leads to even more expression of that gene, etc.).
So what happened in E. coli? By my count 20 out of 26x23=598 combinations (Figure 2a) either failed to be cloned or were cloned but failed to grow. A cloning failure could be due to negative effects on the cell, so presumably only 20/598=3.3% of the reshuffled genes could not be tolerated by the cell. (Note, the authors report this number as 30 or approximately 5%; also note, if you are recalculating this, that there is a control row and column in Figure 2a). Flipping this around 95% to 97% of the rewired plasmids were tolerated by the cell, which frankly is astounding. The authors point out in the introduction their surprise that highly interconnected master switch alterations in the cell can be tolerated.
Okay, so laboratory conditions are easy. The cells are grown under ideal conditions and given everything they need. So, most of the cars started up and are idling in the parking lot; what about taking them out for a test drive? The authors compared growth conditions, of the rewired constructs that were tolerated by the cell only 16% differed significantly from the controls in their growth profiles. 84% of the cars that started up seem to be able to accelerate and cruise normally under highway conditions. (At this point you might start to think that some of the changes made were not significant, like scooting back a car seat a few inches; this is not the case, the authors test the the altered genes are indeed expressed and some of them are expressed at levels 100's of times higher or lower than the controls, and remember these are master switches not randomly selected fine scaled tweaks.)
It's time for a greater challenge; lets take the cars to a racecourse and then off road! The authors did repeated rapid transfer of the bacteria to fresh media---the bacteria have to divide quickly to keep up---and 12 of the rewired networks were able to keep up with the controls and these tended to have rewired flhD controls which regulate flagellar genes and gives a clue as to why this might be an advantage (by suppressing the extra energy it takes to activate the flageller system). Next the authors put the cells under conditions where they either had to survive very long periods of time without fresh media or at high temperatures (50°C, 122°F). They found that a rpoS-ompR rewiring combination out-competed the controls under both of these conditions. So, out of only 598 combinations tried, which is a tiny fraction of the total number possible, one novel combination gave a fitness advantage under a new environment.
This has obvious implications for the adaptation of cells to new challenges by rewiring their gene network. But what is still most surprising to me is how well altered networks are tolerated in general. A cell is much more sophisticated than the machines we are used to like cars. It has evolved to buffer changes and make sure the important things get done despite strong disruptions to the system. Here is another network example, this time from yeast, that might help to illustrate this enhanced level of sophistication compared to our intuition of the system.
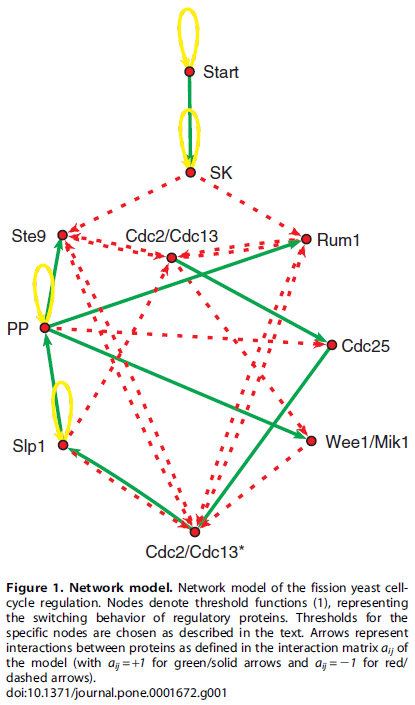
Cells have to undergo a cycle of growth and division. This cell cycle is controlled by a group of genes. In this paper,
Davidich, M. I., & Bornholdt, S. (2008). Boolean Network Model Predicts Cell Cycle Sequence of Fission Yeast, 3(2). doi:10.1371/journal.pone.0001672
the authors treated the cell cycle control genes as being simply "on" or "off" in the following interaction network where the genes turn each other on or off over a series of time steps.
From a starting configuration of gene activity, the start signal (the cell has grown to sufficient size with enough resources) triggers the activity of the other genes flipping each other on and off and a master process unfolds that directs the actions of other genes (outside of the figure) needed to carry out the steps of the cell cycle and division. At the end of the process the original starting configuration is reset to wait for the next start signal (also have a look at Table 2 in the publication). This is a very simplistic model but it captures essential components of what is known about the yeast cell cycle.
What if the starting configuration is disrupted? Then the wrong cascade of signals would propagate through the network, activating the wrong sets of genes. Without a master record of which switches should be set to on and off at the beginning is the cell doomed to deviate along a different path and not be able to return to appropriate cell cycle? Treating the genes (and the start signal) as simply on or off there are 1024 possible starting configurations. This plot shows how all possible configurations are predicted to transition to and from each other.
The arrows in blue are the normal steps of the cell cycle. From a large number of deviated starting configurations the cell will be able to, within a few steps, predominantly reset itself to the correct cell cycle. This is a property of the network of gene interactions and is not due to random chance. (There are some starting points that do not return to the main path, but also keep in mind that this is a very simplistic model.) This shows that the cell has evolved to be robust to disruptions, even in very subtle ways that may not be obvious at first, such as the wiring of its gene interaction network. Simply looking at Figure 1 above does not imply, to a human, the robustness of the system that is uncovered in Figure 2.
The paper goes on to describe another example of the evolution of gene networks with a different set of interactions of the genes involved in the cell cycle for a different species of yeast (S. cerevisiae vs. S. pombe). The wiring is altered, with a different type of reliance on internal signals, but the end result of robustness of the system is essentially the same.