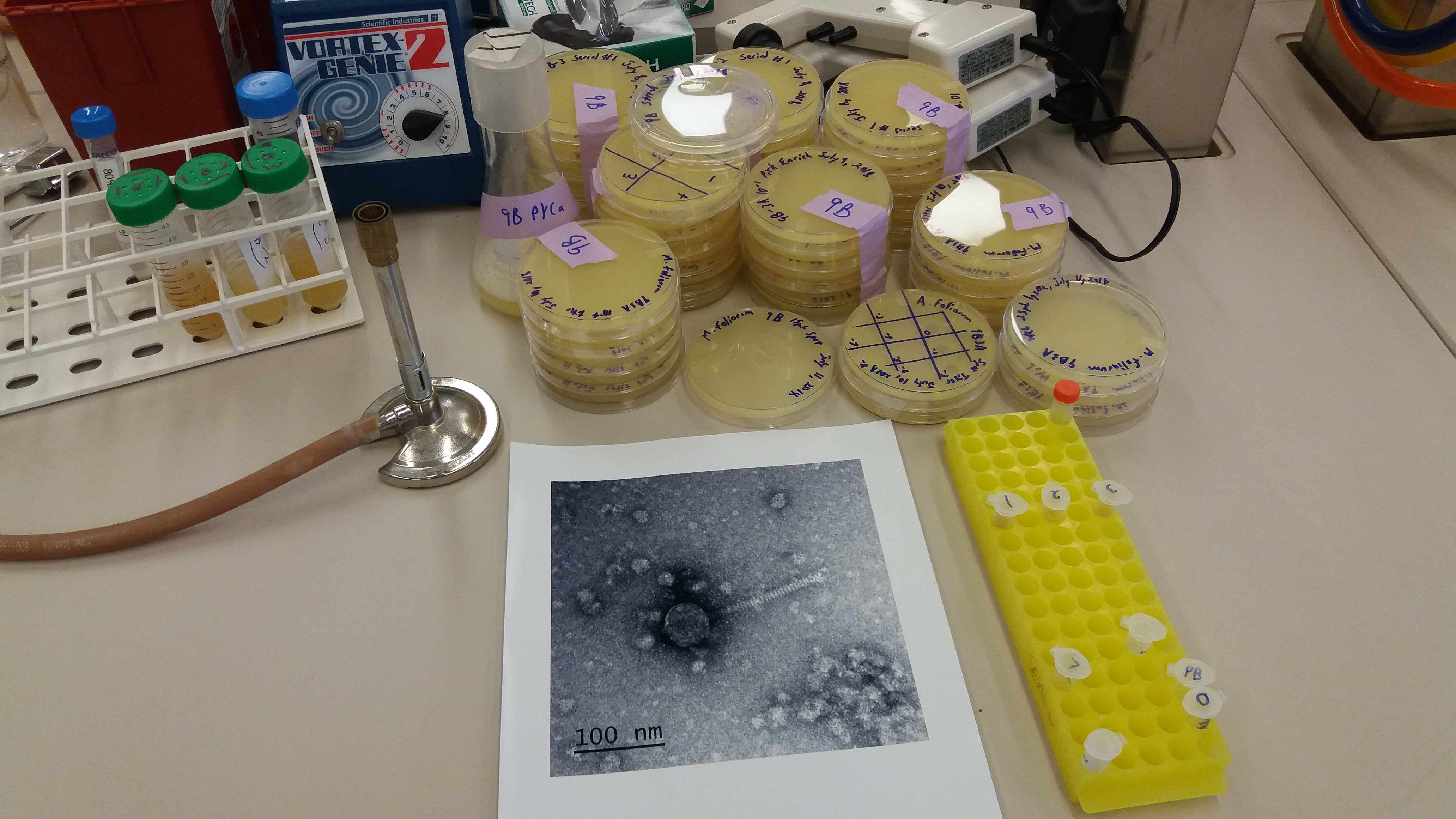
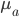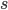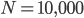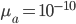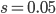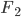I just returned from the 2018 AGA symposium organized by Rosemary Gillespie in Waimea. It truly was one of the best meetings I have been to. The talks and posters were excellent and it was a good group of people to talk to and catch up with. There was a large Hawaiian delegation but also people from all over the world.
The meeting was organized around the questions of adaptive radiations, of which Hawaiʻi has several excellent examples including the Hawaiian honeycreepers, Hawaiian Drosophila, Tetragnatha spiders, Cosmopterigidae fancy case caterpillars, Hawaiian Bidens, Hawaiian lobelioids, ‘opihi, Hawaiian crickets, and silverswords. There were also examples from outside Hawaiʻi such as the Galápagos finches, African cichlids, Aquilegia, stickleback, and Caribbean anoles.
It brought to mind a few things that I have been thinking about on the "back burner" for several years, the inter-island effect and hybridization; however, I am not an expert in theories of speciation and adaptive radiation (especially the more ecological oriented ones related to priority effects and community assembly). A collaborative project on Cottus hybrids (Stemshorn et al. 2011) and learning about the rate of speciation in some groups when first moving to Hawaiʻi started me on this road. Hawaiʻi is very isolated from continents. Once the first two of the large islands formed (Kauaʻi and O‘ahu) I strongly suspect a situation was set up that promoted rapid speciation from intermittent movement between the islands (in addition to other effects related to novel ecological opportunities for newly established species (there is evidence for multiple fitness peaks in an "adaptive landscape" for some species, e.g., Martin and Wainwright 2013 and as evidenced indirectly by convergent multiple ecomorphs, e.g., Muschick et al. 2012 and references therein) in the diverse climates and new species compositions of the islands). A system of parapatry existed where populations could diverge and build (partial) barriers to gene flow yet still remain in contact. There is evidence in the Hawaiian honeycreeper phylogeny that the major speciation events took place after Kauaʻi and O‘ahu (Lerner et al. 2011). It would be interesting to see if this holds up in the timing of other adaptive radiations and the degree of isolation and inter-island distance in other archipelagos to map out the geographic Goldilocks zone for inter-island effects on rates of speciation for different taxa. The other part of this is the role of hybridization (and there is evidence of extensive hybridization in many of these adaptive radiations, e.g., Richards and Martin 2017; Jay et al. 2018; Poelstra et al. 2018; see also Pfennig 2007 for a natural case of adaptive hybridization). Hybridization between closely related species breaks a lot of the standard rules and assumptions in population genetics related to the rate of adaptation. We usually think of a new rare allele resulting from a single mutation that is adaptive. It might become established in the population but in fact, counter-intuitively, it is overwhelmingly likely to be lost to drift (despite being adaptive) in the first few generations. We can write down predictions for the rate of adaptive evolution from population genetic theory like 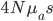 , where
, where  is the population size,
is the population size, 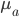 is the adaptive mutation rate (let's say per gene or group of genes), and
is the adaptive mutation rate (let's say per gene or group of genes), and 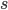 is the relative fitness advantage (see Patwa and Wahl 2008 for review). Rough guess values like
is the relative fitness advantage (see Patwa and Wahl 2008 for review). Rough guess values like 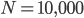 ,
, 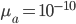 and
and 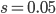 gives an average rate of a genetic adaptation event once every 5,000,000 generations. Long story short, the rate of adaptation is predicted to be painfully slow, especially in small populations such as those that might be found on remote islands. However, hybridization doesn't work like this. In parapatry species hybrids can be constantly generated and the alleles are (so long as the parental species do not go extinct) never lost by drift. Thus it is essentially certain that adaptive differences will become established (giving a evolutionary boost over their single homogeneous species relatives on the mainland). Even better, these are variants that have been pre-tested in the parental species to work well in certain genomic backgrounds rather than new mutations that are more likely to either be unconditionally neutral or deleterious. And, once the process proceeds to the
gives an average rate of a genetic adaptation event once every 5,000,000 generations. Long story short, the rate of adaptation is predicted to be painfully slow, especially in small populations such as those that might be found on remote islands. However, hybridization doesn't work like this. In parapatry species hybrids can be constantly generated and the alleles are (so long as the parental species do not go extinct) never lost by drift. Thus it is essentially certain that adaptive differences will become established (giving a evolutionary boost over their single homogeneous species relatives on the mainland). Even better, these are variants that have been pre-tested in the parental species to work well in certain genomic backgrounds rather than new mutations that are more likely to either be unconditionally neutral or deleterious. And, once the process proceeds to the 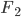 generation a huge diversity of combinations appear for selection to act upon (although the majority of rare hybrid offspring will be back-crosses to one of the parental species) and explore variants that may be near neutral in the parental configuration but novelly adaptive when they interact. Also, note that as two species diverge, assuming that they proceed to complete genetic isolation, they will transition through the full range of levels of divergence up until the point hybrids are sterile or inviable, testing the widest range of levels of genetic differentiation possible. As new species become established, beyond the first two that may be largely allopatric, (and as new islands form) the range of potential crosses and combinations is also increased.
generation a huge diversity of combinations appear for selection to act upon (although the majority of rare hybrid offspring will be back-crosses to one of the parental species) and explore variants that may be near neutral in the parental configuration but novelly adaptive when they interact. Also, note that as two species diverge, assuming that they proceed to complete genetic isolation, they will transition through the full range of levels of divergence up until the point hybrids are sterile or inviable, testing the widest range of levels of genetic differentiation possible. As new species become established, beyond the first two that may be largely allopatric, (and as new islands form) the range of potential crosses and combinations is also increased.
What I am focusing on here is a positive feedback system where, once the initial conditions are met, there could be a runaway increase in diversity, niche exploration, subdivision, and species generation. There are also forces that would work against this such as stochastic extinction of rare species as environmental niches become more finely subdivided, extinction by hybridization, and the inability to evolve boundaries to gene flow of novel types (incidentally influences upon mating systems are also implicated as important components of adaptive radiations, e.g., Ellison et al. 2011; Wagner et al. 2012 see also Roberts et al. 2009 for an unusual example of two simultaneous sex determining systems). These processes could be modeled to run some simulations as well as analytically explore this. However, there are large unknowns to work with. In this situation the largest is probably the distribution of potential fitnesses, mapped to potential niches, of species hybrids coupled with the degree of divergence and genetic isolation of the parental species as well as the potential of hybrids to evolve isolation—which might suggest areas to focus research and simplifying assumptions can be made to make the system tractable.
(And, as a final disclaimer, again, I am not an expert in this field so there might be a lot of literature that discusses these very ideas that I am not aware of.)