We found a total of five Toxorhynchites larvae in a pool containing Culex larvae near Manoa stream (Jan 18, 2018, 21.299882, -157.813210). I haven't seen this genus before; they are possibly T. splendens. These are unusual mosquitoes in that they do not blood feed. The larvae eat aquatic invertebrates including other mosquito larvae and several species were introduced into Hawaiʻi as an attempt at biocontrol of mosquitoes. They are also very large mosquitoes and the genus includes the largest mosquito species. Thanks to Matt Medeiros and Jessica Mavica for helping me ID them.
Monthly Archives: January 2018
Jessica Mavica, Pirbright Institute
Jessica Mavica visited our lab last week and gave a presentation in our department seminar series. She works at the Pirbright Institute (https://www.pirbright.ac.uk/) and took some Culex mosquito larvae back to the institute for research projects there.
Transcriptional effects of a positive feedback circuit in Drosophila melanogaster
A new paper is out at BMC Genomics; it was published a few days before the new year.
Transcriptional effects of a positive feedback circuit in Drosophila melanogaster
This is work that was started many years ago. The tTAV gene expression system was developed into a genetic pest management tool by various groups (e.g., Horn C, Wimmer EA. A transgene-based, embryo-specific lethality system for insect pest management. Nat Biotechnol. 2002;21:64–70; Alphey, Luke. "Expression system for insect pest control." U.S. Patent 9,121,036, issued September 1, 2015). A positive feedback form of the tTAV system was being marketed by Oxitec as "RIDL." In this system a transgene promotes its own expression by binding to DNA. If left unchecked this causes a runaway feedback loop and is toxic to the organism. However, the protein produced also binds to tetracycline and if tetracycline is added to the insect's diet then the lethal effect is masked (tetracycline bound protein does not bind to DNA and drive gene expression).
In a nutshell, in applications large amounts of insects can be reared with tetracycline. Then released into the wild to mate with wild individuals. The toxic effect is dominant and kills off the offspring of the wild individuals that inherit it (who presumably do not have tetracycline in their diet). If enough individuals can be released relative to the wild population this can cause a reduction in wild population numbers over the following generations. This is similar to classical SIT (Sterile Insect Technique) for population suppression.
We were curious about testing this system in various ways. In the first exploration we added tetracycline to food for fruit flies to see if there was an effect. Tetracycline can disrupt normal mitochondrial function and kill naturally occurring bacteria including endosymbionts like Wolbachia. This was the basis of a student's research project: Müller, H., Krefft, M., Reeves, G., & Reed, F. A. 2010. Trans-generational influence of tetracycline on Drosophila melanogaster (Bachelor Thesis, Fachhochschule Bingen). He found a reduction in egg laying and a delay in development in response to tetracycline. Interestingly there was a transgenerational effect in both the paternal and maternal sides (maternal could be due to mitochondrial or Wolbachia effects, but these do not explain the paternal effect).
Next we engineered the tTAV system in Drosophila melanogaster and performed an EMS (ethyl methanesulfonate) mutation screen to see if any mutations could result in tolerance of the feedback loop and survival off of dietary tetracycline. This was the basis of a grant application: Deutshe Forschungsgemeinschaft (DFG , German National Research Foundation). Die Entstehung von Resistenzen gegen genetisch induzierte Sterilität bei Insekten. (The evolution of resistance to genetically induced sterility in insects.) 2010 RE-3062/2-1. Long story short we generated ~400,000 embryos from flies exposed to EMS and recovered five individuals that had the tTAV insert but could survive without tetracycline. All five were male. We tried mating them to females to recover offspring---and all five appeared to be sterile which essentially ended that experiment.
In this paper we wanted to determine the genome-wide transcriptional response to the runaway tTAV effect positive feedback loop.
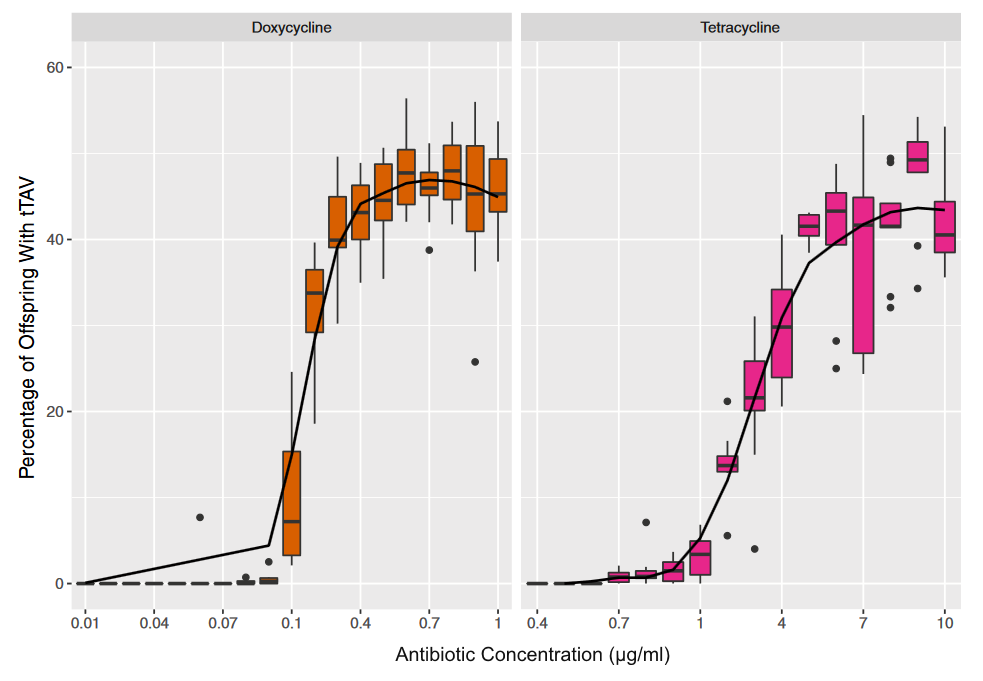
The tTAV insert works as expected in D. melanogaster. The image above shows the curve of lethality as the concentration of two antibiotics, doxycycline and tertracycline, declines. At the upper end of the curve approximately half of the offspring of a heterozygous parent carry the genetic modification, as expected. As the dietary concentration diminishes the curve approaches zero---none of the offspring that carry the tTAV insert survive.
The cause of this lethality is unknown. One can imagine that a positive feedback loop of gene expression and protein production may over-use transcriptional and translational resources and/or overload protein degradation pathways, or disrupt off target gene expression. These hypotheses are not mutually exclusive. Looking at the shift in expression of other genes in the genome in response to tTAV runaway expression could be informative and give us clues as to what process results in lethality.
While we did see thousands of genes across the Drosophila genome that significantly changed expression, there was no clear pattern that emerged. The tTAV system was inserted at different points in the genome and these different lines gave different groups of genes with altered expression. A relative handful of 31 genes with differences in expression were shared across all lines. However, 27 of these 31 genes shifted in expression in different directions, some with increased expression and some with decreased expression across the lines.
This left just four genes with consistent changes in expression when the tTAV system was activated: crok (CG17218), Cyp6a17 (CG10241), olf186-F (CG11430), and Pex23 (CG32226). There is no clear relationship between these genes nor any obvious hypothesis that presents itself in terms of understanding a response to shifts in tTAV activity.
We backup up and looked at gene ontology enrichment. Were there categories of genes involved in certain processes or pathways that tended to be represented in the shift of gene expressions associated with tTAV---again no clear pattern presented itself.
Were there regional changes in expression, perhaps related to different insert sites or nuclear architecture interactions among chromosomes? No, we looked at groups of genes in sliding windows along the genome and there was no clear pattern of clustering by region.
Keep in mind however, there were widespread and significant changes in gene expression. It was not that there was the lack of an effect; just that there is not a clear pattern to the effect.
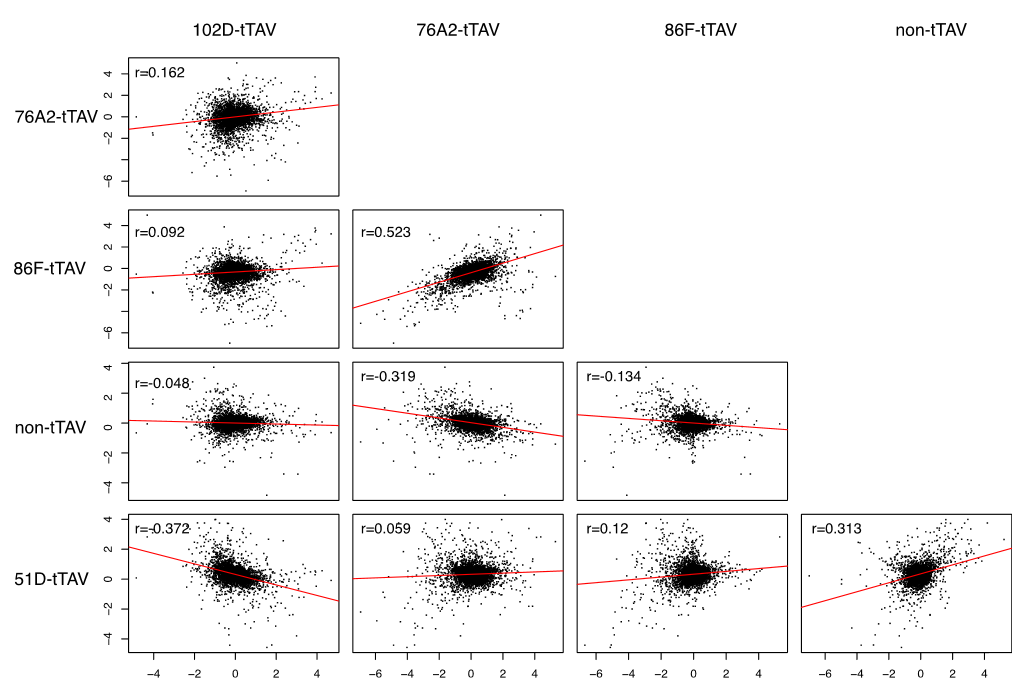
Gene expression levels between pairwise comparisons of the different insert lines and the control all showed statistically significant correlations. However, the direction of the correlation was not consistent. (This is shown in the figure above.)
I was quite surprised by this result---that the insertion site of a highly expressed transgene has such a profoundly idiosyncratic effect on genome-wide transcription patterns. This was a bit frustrating to be honest but it does suggest some cautions in these kinds of studies and next steps to explore. First of all a a caution, if we had only studied the transcriptional response to a single insert we would have found a significant response, with many genes affected, and may have been misled by the resulting pattern (if for example a number of these genes were in a certain pathway), overgeneralizing the particular signal seen. In a next step, it would be interesting to see if this widespread idiosyncratic effect extends to other genetic modifications or is specific to the tTAV system. For example, what is the genome-wide translational response, if any, to actin5c driven expression of EGFP? Importantly, does any effect seen vary widely by insertion site?
Gene Drive Questions
I received an inquiry about gene drive technology by email and thought that it might be useful to post part of it along with my reply here.
Here is the question:
"Since a few months, gene drive is a very prominent topic. But what I am missing a recent review about the state of the art in building gene drive constructs and testing them. I would like to ask you if you could provide some advice about literature, which would cover an overview about this theme. At the level where I try to follow the discussions, I have the feeling that things are randomly mixed (like e.g. gene drive = synthetic biology). It would be really great to have an overview about what is possible by now, what has been done, which systems work and which do not work. And perhaps where is gene drive applied."
And my response:
This is rapidly developing. A lot of the recent focus is on CRISPR-Cas9 based gene drive and there is some confusion in the media that this is the only form of gene drive. I've tried to categorize the different forms in Appendix A of this article:
One major problem with CRISPR-Cas9 based gene drive is the rapid accumulation of mutation and naturally occurring resistance in the wild. See:
http://www.genetics.org/conten
http://www.genetics.org/conten
http://journals.plos.org/plosg
https://www.biorxiv.org/conten
Work is also progressing on underdominance-type approaches that may get around some of the problems with the CRISPR system.
The Genetic Engineering and Society Center at NCSU has a lot of resources and knowledgeable people in this area.
Here is a series of recent publications that were recently posted---I haven't had a chance to go through all of them yet.
https://research.ncsu.edu/ges/
There are also some relevant publications that will come out over the next few months. I can update you as these are published and become publicly available.
To my knowledge there is no direct application as of this moment, but this is likely only a matter of time.
The US military is also very interested in gene drive issues. DARPA is funding some of the research on gene drive and IARPA is working on ways to detect if genetic modifications have occurred.
https://www.darpa.mil/program/
https://www.iarpa.gov/index.ph
Let me know if I can help with additional information or if you want more detail about something.