The Polymerase Chain Reaction (PCR) is a very powerful technology invented in 1983 by Kary Mullis. We use it routinely today in molecular genetics labs and it is easy to take it for granted. What makes this reaction particularly significant is not only that it uses a enzyme catalyst that remains unchanged while driving the reaction, but also that it results in an exponential amplification of a defined DNA sequence. Human intuition tends to underestimate the power of exponential amplifications. Casually imagining that you get paid a penny on the first day of a job and that your pay doubles each day (two pennies on the second day, four pennies on the third day) seems like it would take a long time to make much money--at the end of the week you are only up to $1.28. However, you would have well over 10 million dollars (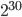 pennies) by the end of the month! Kary Mullis was only given a $10,000 bonus for inventing PCR by Cetus, the company he worked for; but in 1993 he was finally awarded the nobel prize in chemistry for his invention.
pennies) by the end of the month! Kary Mullis was only given a $10,000 bonus for inventing PCR by Cetus, the company he worked for; but in 1993 he was finally awarded the nobel prize in chemistry for his invention.
PCR works like this on DNA. A stretch of DNA is designated with a set of primers (short single stranded DNA segments that can be synthesized). A DNA polymerase enzyme attaches to one end of the primers and synthesizes a new DNA strand in one direction matching the original template DNA. There are cycles of heating ("melting" the DNA by breaking hydrogen bonds between the strands), annealing of the primers to single stranded DNA, and extension of the new strands by the polymerase. Importantly, the newly synthesized DNA strand can be used as a template in the next round, so, in theory, the amount of DNA between the primers is doubled each round. If you do this for 30, 35, or even 40 cycles, you can amplify enough DNA to work with and sequence from even a single starting molecule.
Here in Hawai'i there is currently a GMO food labeling debate. Unlike the EU, crops that are genetically modified do not have to be indicated on food labels in the US. This fits into the larger GMO crop debate and is something that the students in my class are very interested in learning about. In fact, just after I arrived here in August 2011 there were newspaper headlines about acres of transgenic crops destroyed on the big island and rewards for information (link). This led me to thinking about a lab activity that would capture their interest--detecting GMO food ingredients by PCR.
Almost all GMO crops use the same strong constitutive promoter (highly expressed and always on because of a high affinity for RNA polymerase) from the cauliflower mosaic virus (CMV35S) to drive expression of the inserted gene and a "nos" (from nopaline synthase) terminator from the Agrobacterium tumefaciens Ti plasmid to stop transcription of the gene on the opposite side. Primers to detect these parts of the genetic insert are already published so I ordered some to try out.
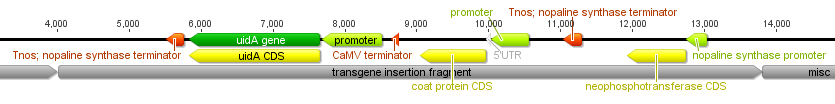
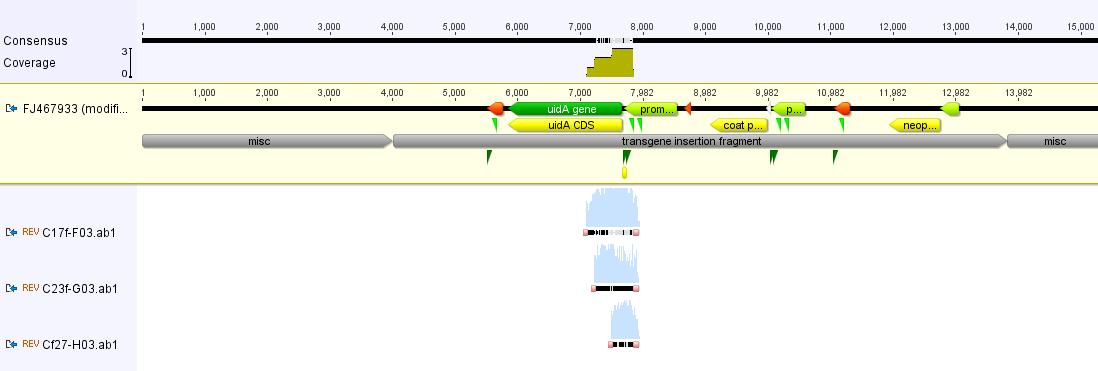
The image below is an overview of a 9+ kbp (over nine thousand base pairs) genetic modification inserted into transgenic papayas to combat the papaya ringspot virus (the virus devastated the papaya industry here in Hawai'i over the last 50 years). The type of insert varies for each application, but as you will see later on, there is a reason I picked the GM papaya to illustrate.
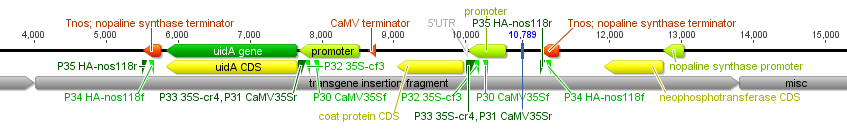
And below I've indicated the positions where three sets of primers are predicted to anneal: one set in the nos terminator and two sets in the CMV promoter indicated by the green triangles.
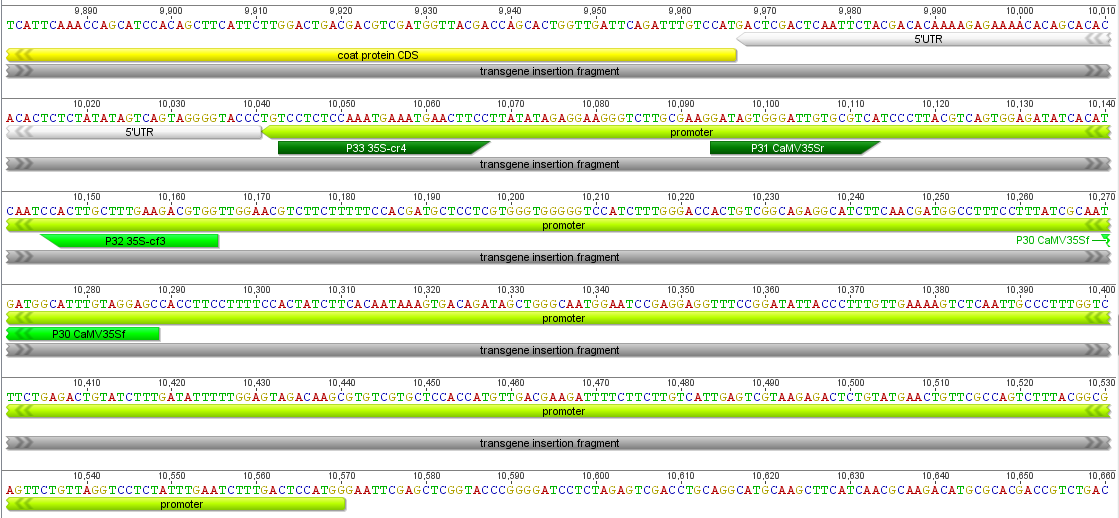
Below I have zoomed in a bit to see more detail; the sequence is so large it has to be wrapped on the screen so moving off to the right comes back on the next row down on the left. In light green promoters are indicated, protein coding genes are in yellow and terminator sequences in orange.
Below is a closer picture of part of the CMV promoter with "forward" and "reverse" primers. This promoter is used to drive expression of a coat protein from the ringspot virus, to prime the papaya's immune response to the virus--this particular genetic modification is often credited with saving the papaya industry here in Hawai'i. The particular primer pair I tried as of this moment was the nos pair and one pair in the CMV promoter; I have not tried the second CMV pair yet.
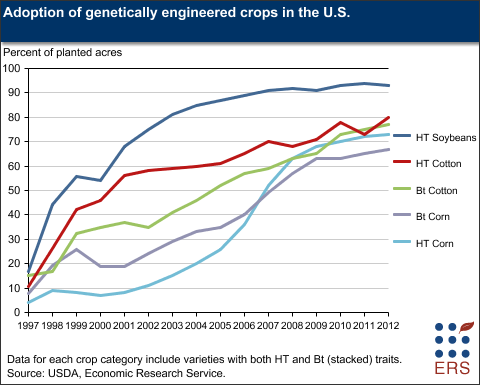
Now it's time to go out and get some samples to try them out on. In the US the big three are soybeans, corn and cotton (the image below is from here).
I couldn't think of a source of cotton with enough DNA to easily test, so I focused on corn, soybeans and--here in Hawai'i--papaya.
At the local grocery store I found "unlabeled" corn from the US:
"Green Giant Nibblers" also grown in the US:
And "GloriAnn Sweet Corn" a product of Mexico:
The only soybeans I found were all products of China. "Shirakiku Edamame":
"WelPac Shelled Edamame":
And "Safeway Kitchens Shelled Edamame":
For Papayas I found "Diamond Head Papaya":
Anonymous "Papaya" from a different store:
And "Solo" variety papaya from a farmer's market:
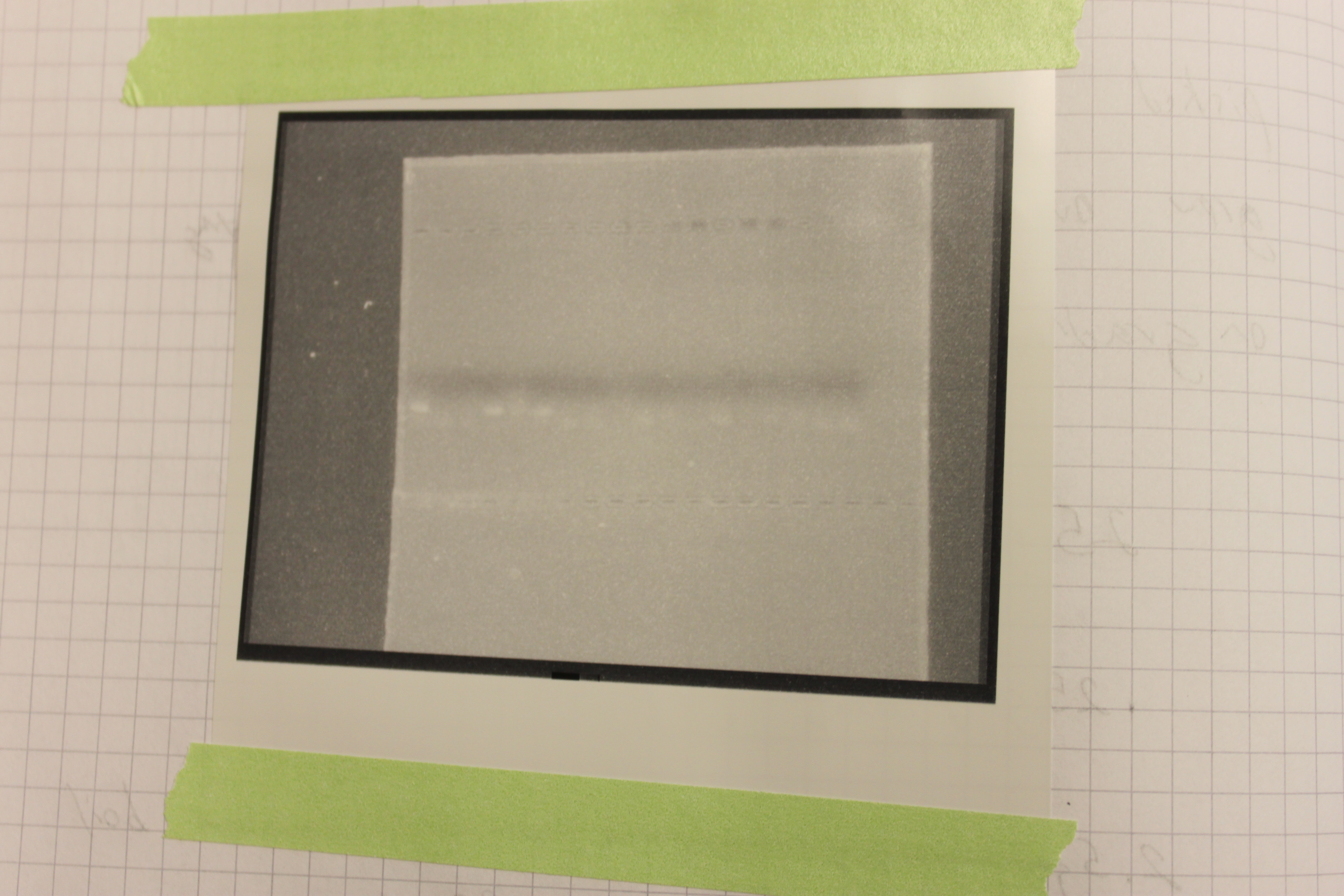
So we have three of each of corn, soybean and papaya samples. After extracting DNA samples, running the PCR and running a gel to view the results, this is what I found:
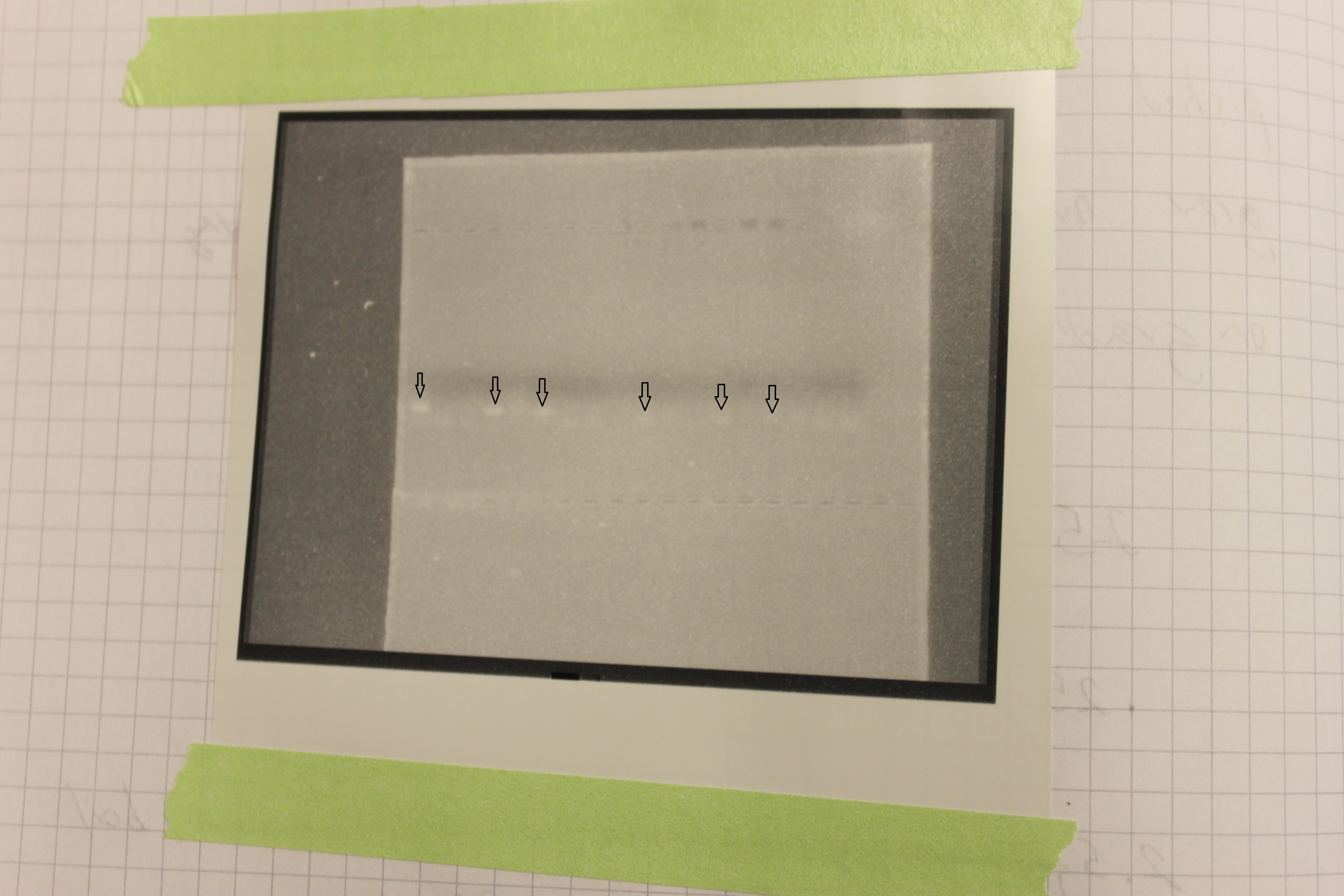
To the left are the CMV primers and to the right the NOS primer pairs. Three lanes in each showed up positive for amplification of the corresponding DNA. The positive bands are faint so I indicated them with arrows in the image below.
These primers amplify a short DNA sequence which does not stain with much dye to see on the gel. The CMV promoter primers worked "better" i.e. are easier to see. If you look closely you can also see a fainter band just below the positive samples that is "primer dimer". This happens when the primers anneal to each other and amplify an even shorter sequence from just the primers. Primers are designed in pairs to minimize this effect however.
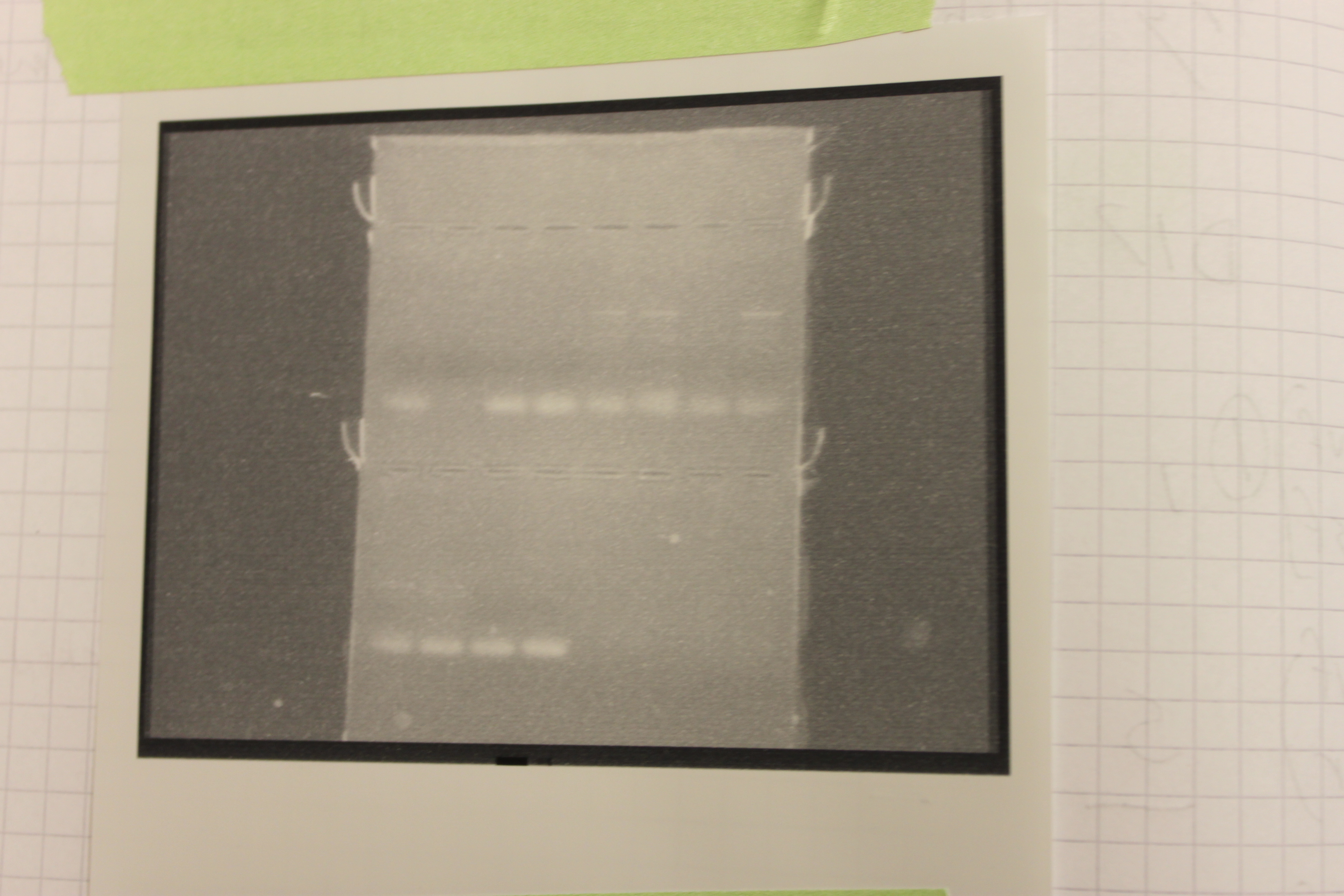
In the gel below I tried different combinations of the CMV and NOS primers together in the same PCR reaction to try to amplify the larger sequence of DNA between them. In this case these primers were not designed to work together and the primer dimer is much stronger at the bottom of the lanes. There are three lanes that gave a large (did not move as far down the gel) band in the upper right (CMVf, NOSr). The PCR in the second lane in from the top left didn't seem to work at all, this could be due to a PCR inhibitor that was co-extracted with the DNA.
So what were the three positive samples? They were the three papaya samples, two from the store and one from the farmer's market. To nail this down a bit more I sequenced the DNA that was amplified to make sure it is what I suspected.
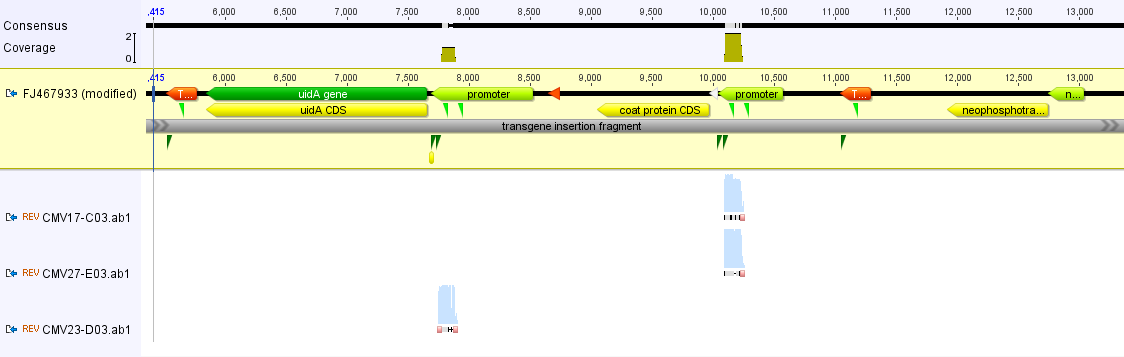
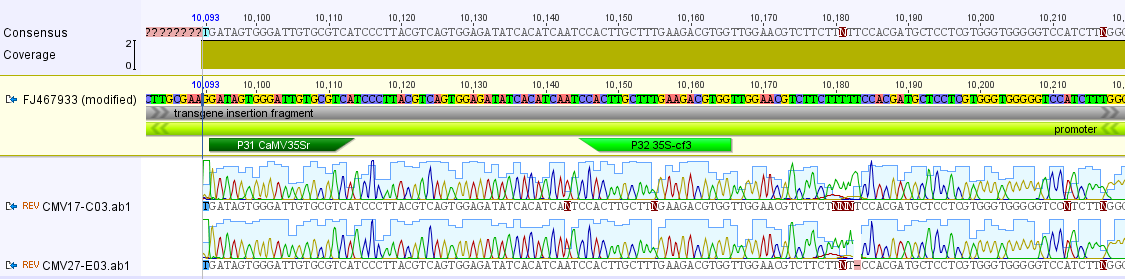
Above, the short sequences from the three samples (gray bands in the lower part) are mapped to the papaya insert as a reference sequence. As expected they map to the CMV promoter. Below I zoomed in some more on a section so you can see the DNA sequence, it is a match to the promoter sequence.
Above, two matches to the "right" promoter and below one match to the "left" promoter.
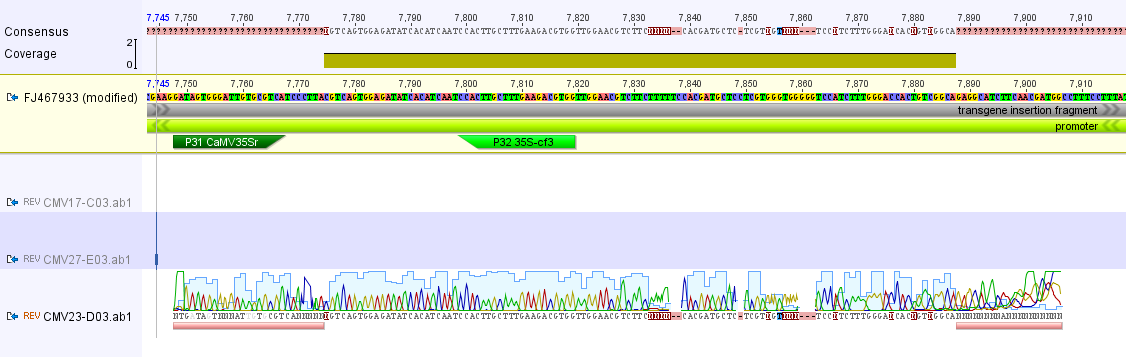
Below is the longer sequence I recovered from the CMV-NOS primer pairs.
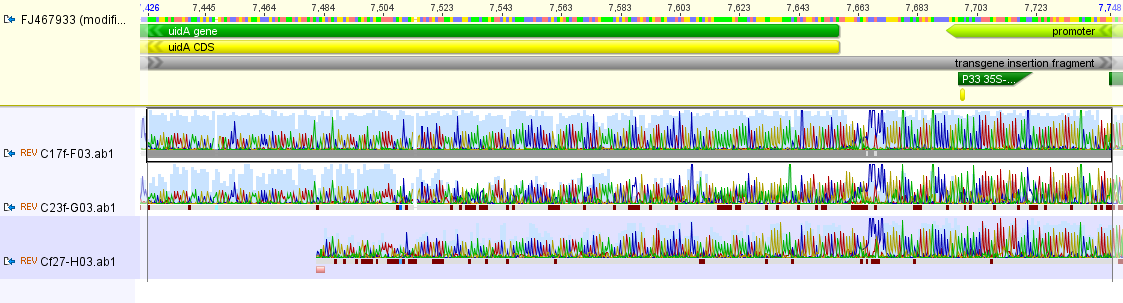
Zooming in more below you can see (hopefully) it is a match to the uidA gene in the genetic modification insert. (The colors in the trace file correspond to the different colored bases in the reference sequence at the top.)
uidA is a gene from E. coli and produces the enzyme beta-D-Glucuronidase. When plant tissues are treated with a bromine containing compound (X-Gluc) that is cleaved by beta-D-Glucuronidase and stains the tissues blue. This provides visual confirmation that the gene in the insert is being expressed (similar to the X-Gal reporter system in E. coli) and is called the GUS reporter system. The point being here, this is not a gene sequence (with components originating from a virus, CMV, and two different bacteria, NOS and uidA) that would be present in non-genetically modified papaya, but is exactly what we expect in ringspot virus resistant genetically modified papaya.
Let me back up a moment at this point and put a disclaimer in here. I did this informally in the lab to test for use as a teaching tool in my genetics class in the fall. There is a chance of a false positive here from DNA contamination between samples. As I said at the beginning, PCR is so powerful it can amplify a single DNA molecule so this is something to always keep in mind. There are ways to address this with multiple independent extractions in different labs, amplification from different parts of the insert between different samples, looking for unique genetic differences between the samples (if they exist), etc. but I am not going to go into that detail here.
Also, while we are on a cautionary topic, the CMV promoter is from the cauliflower mosaic virus. It is possible that cauliflower and related brassica may show up positive even if they are not genetically modified from natural CMV infection. Agrobacterium infects certain plants naturally and even E. coli can contaminate fruits and vegetables from contaminated water sources (which can be a serious health problem). But you would not expect to find the CMV - uidA combination above. That is proof that these papaya (or at least one if we are being hyper-cautious) are transgenic genetically modified.
I was surprised that the "solo" variety from the farmer's market showed up positive for the ringspot virus insert. This is not supposed to be a genetically modified variety. However, if you do a search for the terms "papaya gmo seed contaminated" you can find that it is quite common for papaya varieties, solo included, to contain the genetic modification here in Hawai'i, unknown to the growers, sellers and buyers using them.