We met with Prof. Scott O'Neill and his lab today. We discussed what we are trying to do in Hawai'i with underdominance and possibilities with Wolbachia in mosquitoes in Hawai'i and Wolbachia strains from Hawaiian Drosophila species. He is dean of the college and very busy but took time to talk with us and set up a tour and meetings with people working in his lab. Their main focus is on modifying mosquitoes to block the spread and transmission of Dengue (the virus that causes Dengue fever) by using Wolbachia injections. Long story short, certain Wolbachia (a type of symbiotic bacteria) strains can infect and be stably transmitted over generations in mosquitoes (and many other insect species). If at a high enough starting frequency they increase in frequency in local populations and move toward 100% frequency. Importantly, Wolbachia has also been shown to reduce the transmission of certain disease causing viruses and Plasmodium. Originally they used a MelPop strain (from Drosophila melanogaster fruit flies) that reduced mosquito lifespan and virus transmission but it reduced fitness of the mosquito too much in the wild and would be lost. They switched to a Mel strain (also from Drosophila melanogaster) that also (incompletely) blocks transmission and is able to become stably established in the wild. They are conducting releases of Wolbachia infected mosquitoes in Australia and work in SE Asia. We also discussed the regulatory aspects of their work both in Australia and internationally. We got to see how they were raising and working with the mosquitoes, from cages to collecting and feeding to biosecurity setup (most of the work is at level 2 but they have a level 3 lab for the Dengue virus work). We even got to see them do mosquito (Aedes aegypti) egg micro-injection and Jolene had a shot at arranging the eggs under a microscope and injecting them with a new experimental Wolbachia strain using a micromanipulator. The people here were extremely helpful and friendly and we got to ask them questions for several hours---we were literally there all day and they took us out to lunch midday. We have plenty of new pointers to help us with working with the mosquitoes in our lab. The data they are getting back from the releases are impressive. They are collecting mosquitoes from the release sites and areas around them weekly and plotting maps of the increase in allele frequency of the Wolbachia infected mosquitoes and the geographic spread from the release range. On top of this you can see population fluctuations, drift, the effects of rainfall on the wild populations, etc.
Category Archives: Uncategorized
We're in Australia!
Jolene and I are off to Australia. First to meet with Prof. Scott O'Neill at Monash University in Melbourne then to the 2014 GSA (Genetics Society of Australasia ) conference at the University of Sydney.
A nature walk
I had some free time after the conference ended and before my flight back to Hawai'i---this is very rare for me. So, I took advantage of the opportunity to go for a nature walk around the UMBC campus. I saw lots of plants and animals including several deer, rabbits, squirrels, a groundhog, geese, barn swallows, etc. I also brought along a camera and got photos! This is a space holder post for now, because I have to get get ready to the airport soon, but I will update with some pictures and commentary as soon as I get a chance.
Synthetic Biology Workshop, Done
The HHMI/NSF sponsored workshop on using synthetic biology as an undergraduate teaching tool has finished. I am really glad I came to this. I learned about some impressive new tools, Golden Gate Assembly, GoldenBraid Cloning, C-dogs for uniform gene expression control, reasons to use doubled translation termination sites in plasmid design, The Oligator, new long strand cheap oligo synthesis technologies, just to name a few. And a lot of good ideas to use both in teaching and research. We put together 10 minute presentations that are proposals for ways we might use synthetic biology teaching at our home institutions and presented them today. I teamed up with Mark Wilson of Humboldt State University and we presented something about using Vibrio bacteria as a marine model in synthetic biology. I have already uploaded a version (edited for clarity---this is not the cleanest presentation I have made but go easy on me---it was put together quickly in a very short amount of time as a part of the workshop) of our presentation to the posted presentations page.
There were also some other impressive presentations. One that I really liked was a way to use plasmid construction as a random number generator. Generating random numbers is actually quite difficult to do well using software on a computer, but it may turn out to be easy using the orientation of a promoter with two reporters and a FACs machine (fluorescent activated cell sorter)! The same group also suggested using a mix of transformed bacteria colonies to simulate genetic drift and/or selection with repeated sampling and plating. There was another presentation on using E. coli to produce wintergreen to prevent colony collapse disorder (due to mites) in honeybees. And also a presentation on designing bacteria for rapid, efficient, and specific gun residue detection at crime scenes, etc.
Synthetic Biology Workshop
This week I am on the US East Coast for a GCAT synthetic biology workshop sponsored by NSF and HHMI. The focus of the workshop is on using synthetic biology as a tool to teach undergraduate genetics research. Naturally I was speculative about just how "new" synthetic biology is, etc. but I am already impressed by some of the things we went over at our first sessions last night: the Golden Gate plasmid assembly process for example. Some of the focus of synthetic biology (which is argued to set it apart from genetic engineering in general) does not come naturally to many biologists but is said to be heavily influenced by engineering and computer science, such as standardization and modularity. There is also a comfort with "black boxes." You don't have to understand the (generally known) details about how something works to use it (indeed the focus is on using any tool available without a detailed explanation of the mechanism), which does not come easy for many geneticists. However, they made some convincing arguments and examples of the utility of this approach. Part of why I am here is to get ideas for undergraduate research and education but I can say I've already seen how this could be useful in plasmid design for some of our other projects.
Also, I am grateful to the conference organizers for breaking some rules to allow me to be here. The workshop is designed to have teams, preferably a biologist and non-biologist, come from an institute to be trained and take back cross disciplinary ideas with them to their home institute. I could not find anyone at UH to join my team before the application deadline; so I figured I had nothing to loose and sent in my application anyway with a brief explanation, and was accepted solo!
Frustrating Review Process
At times the peer-review process can be very frustrating. It is probably wiser not to publish this but I feel like I have to speak out about this somehow---if for no other reason than to illustrate part of the publication (and also grant review) process. Our two recent publications were in review at various journals for years. It is painful to see other publications come out that are similar to what you have been trying to publish and to see so much time going by on the calendar during the process. The peer review process is necessary, and on the balance helps improve the quality of scientific publications. I can honestly say that some of my publications have greatly benefited from reviewer feedback. However, this is not always the case. I think removing the authors names and institutions from the material the reviewer sees, among other steps, would improve the process. At any rate, I have saved the following review as an example of what we went through to get our papers published (this one is from a previous submission of the engineering underdominance manuscript). This reviewer followed our manuscript submission from one journal to another (the same person was picked repeatedly by the editors). We could tell this because some of the reviews contained identical word-for-word copy and paste sections despite our revisions to try to address and clarify them. I have added the emphasis but the words are taken directly from the review. If you read through to the end you can see that the reviewer made up speculative scenarios that were not in our paper as a basis to reject it.
August 21, 2012
Unfortunately, the authors have failed to generate the underdominance-based gene drive system they claim to have created, and thus the paper must be rejected. In fact, their text is written in such away that the fact that they have not in fact created a functioning gene drive system only becomes clear when one reads the methods.
In brief, their strategy is to use RNAi, with the dsRNA generated targeting a ribosomal protein, and being expressed under the control of a gal4 uas element. Flies carry this piece of DNA and in addition a rescuing version of the ribosomal protein that is resistant to the dsRNA-dependent RNAi. Flies carrying this chromosome (the third chromosome) are argued as showing gene drive (below). But of course this is nonsense. This chromosome does nothing at all in a wildtype population. Absolutely nothing at all. It does not drive. It does nothing but float in a wildtype population.
The only context in which the transgene-bearing third chromosome does show drive involves a trick, hidden in the methods. The authors take their rnai-rescue line and introduce into this genetic background a transgene on the second chromosome bearing an actin promoter driven version of GAL4, which drives transcription from UAS elements. Thus, in this new strain, acting gal4/Cyo balancer; uas rnai-rescue, we now get effects. The actin-GAL4-driven dsRNA knocks down the expression of the ribosomal protein; in the presence of one copy of the rescue construct (in heterozygotes) they are less fit, and show a lower viability. Very importantly for the authors technique, they are not dead. Otherwise they would never have been able to cross them with each other to generate homozygotes. In homozygotes for the third (The second chromosome balancer prevents GAL4 from ever becoming homozygous), the animals are more healthy than heterozygotes (but of course only heterozygotes that carry a gal4-bearing second chromosome), but less fit than wildtype.
This genetic behavior allows them to show a form of underdominance - in an extremely contrived and artificial situation - that fairly boggles the mind. They take their actin-gal4/cyo; rnai-rescue flies and cross them to their "wildtype" strain, which is balanced on the second chromosome for actin-gal4/cyo but wildtype for the third chromosome. In other words, their wildtype strain always contains actin-gal4, but not in their actual construct. In the text the authors say that the use of gal4 is not essential for their system and is only meant to be illustrative. But this is nonsense. They simply cannot generate drive without the presence of a gal4 that is introduced into their genetic background.
Not surprisingly, in the above, contrived situation the authors can show underdominant behavior. When the frequency of the rnai-rescue chromosome reaches some frequency that depends on the relative fitness costs associated with being heterozygous and homozygous for the chromosome (but only in the presence of a actin-gal4, the benefits of being homozygous for the rnai-rescue chromosome outweigh the benefits of carrying a wildtype chromosome (which at this point finds itself in unfit heterozygotes most of the time) and the uas-rnai-rescue chromosome drives to allele fixation. Below this threshold the chromosome disappears from the population.
Again, the paper must be rejected because they have simply not created a driving underdominant chromosome that can drive itself into a wildtype population. It is remarkable that the authors believe that they can get away without generating a simple three gene construct in a model organism such as drosophila, that would show drive.
In thinking about why the authors might not have done such an obvious and essential experiment one can only conclude that they probably tried but were unable to generate the relevant transformant. There is probably a very good reason for this, and it relates to the reason why no underdominant gene drive systems have ever been created using modern genetic engineering techniques: It is very hard (if not impossible) to generate the initial unfit heterozygote, which will ultimately give rise to the more fit homozygote. Unfortunately, without a demonstrated solution to this problem all engineered underdominance systems are so much hot air. They look good on paper, but in fact cannot be generated in actual living organisms.
For the record, our plan from the beginning was to try different expression patterns of the RNAi "poison" using the GAL4/UAS system to see what expression pattern resulted in the best underdominance properties. The first expression driver we tested, act5c-GAL4, worked so well that we immediately wrote up and submitted our results for publication. To get a single locus system the act5c-GAL4; UAS-RNAi could be substituted with act5c-RNAi to remove the GAL4-UAS middleman. However, this seemed so trivial we didn't consider it important in reporting our first results. The hemizygous (single insert) individuals are viable and fertile and in fact we used them in crosses to generate homozygous individuals with an alternatively marked GFP system. Anyway, tested objective scientific facts seem to fall secondary to dogmatic subjective opinions (see above), even in the scientific literature. How much, from other studies, is modern research missing out on?
The reviewers seem to have wanted us to repeat the experiments in Drosophila with a single insert. However, (a fact of modern science) we have moved on to new job positions in different labs and Drosophila was only used as a convenience to arrive at, in a timely manner, some definitive results. We want to move quickly to our real target, mosquitoes and other species. In the longer view it is a waste of time to back up and try to make the reviewer happy (which is obviously a lost cause from the start). But, unfortunately, reviewers like this are all too common (I am restraining myself from providing additional examples) and delay or prevent getting published and getting necessary funding to continue the research.
And our Medea-Underdominance manuscript is also out in print!
Right on the heels of the last paper our theory results on a combined Medea-Underdominant dynamics is finally published. Like the last one this took several years going from journal to journal and through a long line of reviewers before finally getting published. Chaitanya really deserves the credit on this one for making the final push to resubmit yet again and get it out there.
Our population genetic engineering paper is published!
At long last, our paper describing engineering underdominance in Drosophila with haploinsufficient poison/rescue is out! Here is a link to the full article.
In other news, classes and committee work are finally done, so I can have a chance to get back into the lab. My preproposal to NSF was turned down again but a preproposal to NASA has made it to the next stage to submit a full proposal!
Our DNA Barcoding Project
The basic idea of DNA barcoding is that essentially every species has a unique DNA sequence that is diagnostic of that species. It is inefficient to sequence different genes for comparison of different samples or even whole genomes, so it makes sense to pick a common place in the genome for comparison across species. A great deal of effort has gone into determining the best gene sequence to use for barcoding. It should be variable enough that essentially all species are unique, but not too variable (at least in the primer binding regions) so that the same set of primers can work in almost all species and that reasonable sequence alignments can be made to compare species.
Why? Many species are described at only one stage of their life cycle. In many insects the larvae look alike but the adults are completely different from each other (or in one famous case the opposite is true, link). Similarly, many marine fish have tiny larvae in the plankton that look nothing like the adult forms, and there are cryptic look-alike fish species. There is an organized effort to barcode as many fish species as possible (link). And take the example of mushrooms. The much more common mycelium fibers growing in soil look nothing like their occasional fruit---the mushrooms that we are used to. Also, there is the problem of identifying what unobserved species are in an environment from the DNA they leave behind in the soil or water. In many cases, like monitoring the species in the environment, and understanding which environments are important throughout a species' life-cycle, it would be helpful to be able to connect the dots and to know precisely which described species are present at different developmental stages or just in general.
Also, in a more forensic sense, it is handy to be able to test food and find out which species it comes from. There are laws about which species can be harvested and how food ingredients have to be advertised. DNA barcoding provides a useful and powerful way to verify the claims of a label (e.g. link).
Finally, there is a educationally valuable, direct, tangible connection for the students; a sample they find themselves and decide to select connects to modern molecular biology in the lab and then the results connect to the natural history of evolution on our plant that ultimately connects all living organisms.
However, DNA barcoding has been surprisingly polarizing; it has plenty of lovers and haters in biology. Classical taxonomy is a skill that takes years to develop for a specific group of species of the taxonomist's specialization. Some see DNA barcoding as a cheap and easy way to circumvent this process, resulting in a rapid naming of different species in a sample with no real deeper understanding of the important differences between them. Like any new technological approach there will be an adjustment period as DNA barcoding finds and settles in on its role in biology.
(For more background information about DNA barcoding see the iBOL website, the introduction at CBOL, CSH's barcoding 101, and the DNA barcoding wikipedia page, which includes a discussion of criticisms that are not as forthcoming on the other websites.)
I am teaching a genetics course with a laboratory component (with the first lab last fall semester). In my research lab we started using DNA barcoding to identify which species were present as mosquito larvae in standing water. I expanded this to test some other species I came across and decided to include this as a project for the undergraduate students. In many ways, Hawai'i is one of the most uniquely diverse places on the planet, so describing this diversity capitalizes on a natural strength of Hawai'i.
COI, cytochrome c oxidase I, found in the mitochondrial genome, has, for various reasons, become the standard barcoding locus for animals. Both matk and rbcL have been promoted as barcoding regions in plants and ITS in fungi. We have settled on rbcL for plants because the PCR amplification is more reliable (in our lab experience) over a diverse range of plants than matK. We have not tried fungi samples yet but may include them soon.
Last semester I asked the students to bring in a (legally and safely obtained) tissue sample for DNA barcode testing. We spent some time, effort, and resources on this and I don't want the results to simply disappear at the end of the semester, only existing as a student experience. So, I have been curating the results and adding them to an online database so that researchers and students all over the world can access the results and use them in their own research---including a growing resources for students to compare results to in our teaching lab. This has been somewhat on the back-burner as a free weekend project and I have just now finished submitting the animal samples. There are even more plant samples, especially flowering plants, that I expect to finish curating this summer. So, this post is to talk a bit about the initial results focusing on the animal samples.
Also, in another sense, I am also trying to learn more about the natural history of Hawai'i, and have my own gaps in understanding of species taxonomy---especially in the tropics. Generating DNA barcodes from samples collected here provides a focus to help fill these gaps in. Without this focus, on working out the relationships of the few species at hand, the bigger picture is too overwhelming. This creates a backbone for us to connect new species to as we come across them in the future.
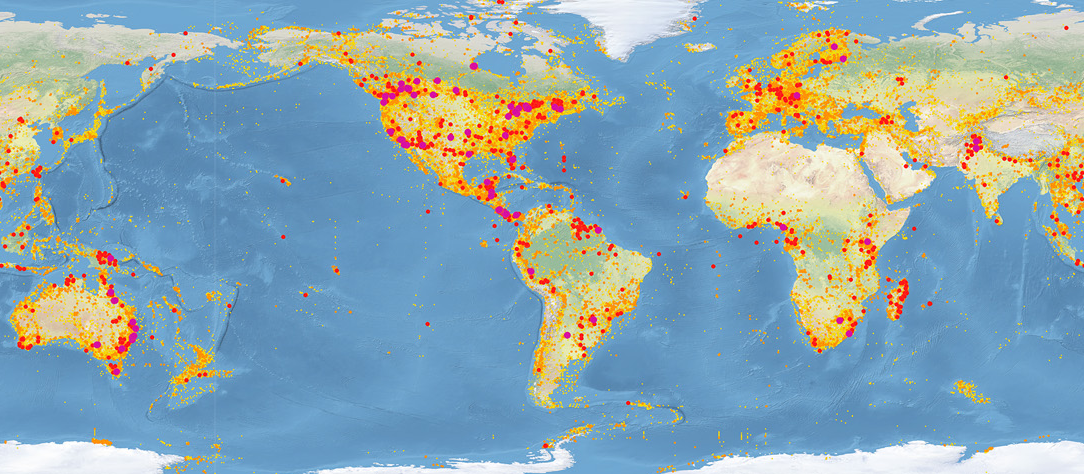
The BOLD Systems (Barcode of Life Data Systems) website is a publicly searchable repository of DNA barcode data. In addition to the sequences you can also record the exact location a sample is collected from, when it was collected, an image of the sample, etc. On their front page there is a map highlighting the geographic distribution of samples in the database.
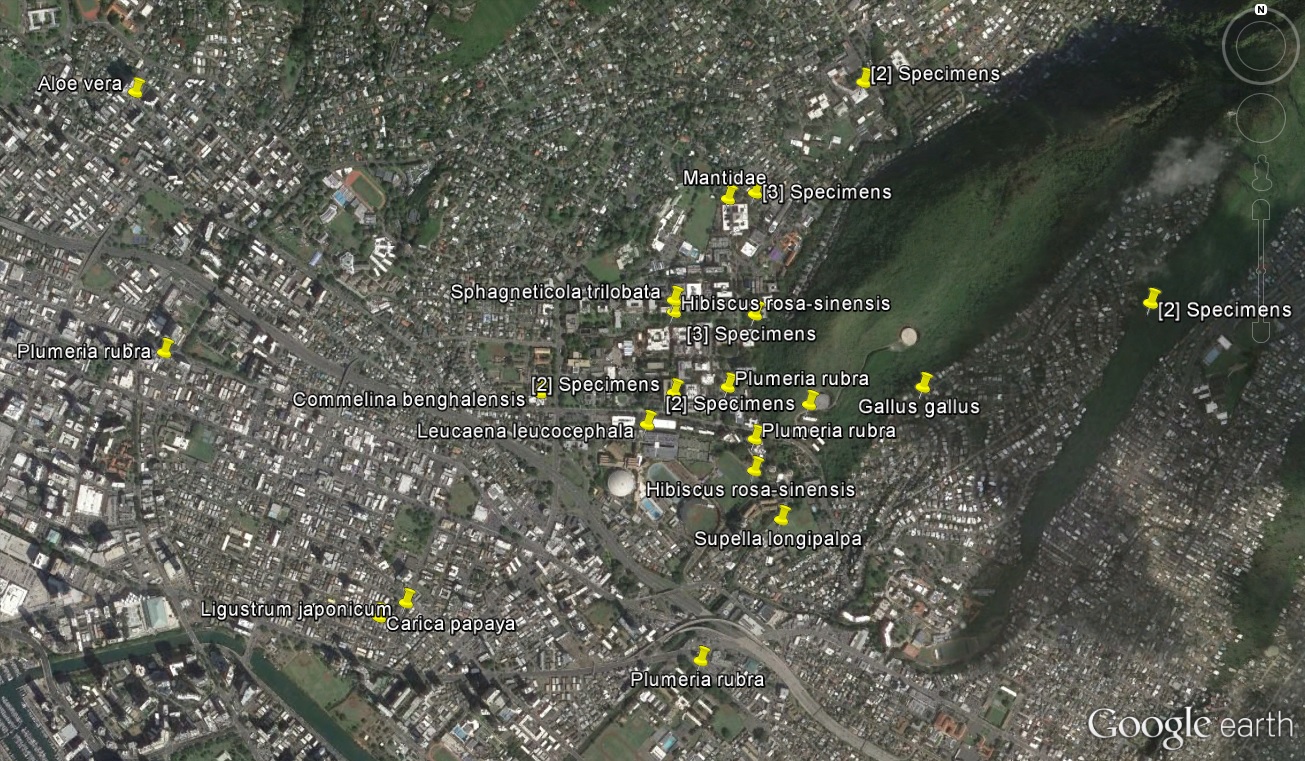
We are contributing to the purple dot over O'ahu in Hawai'i. Zooming in, here is a map of our samples in O'ahu.
And even closer showing the Honolulu/Manoa region around campus.
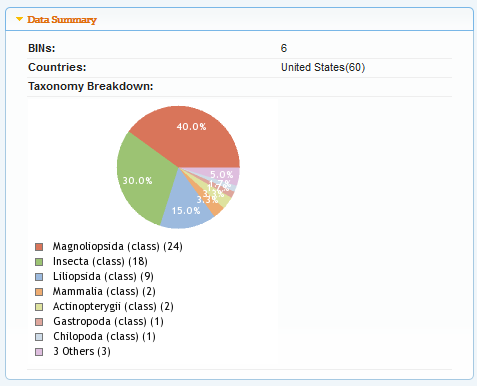
Currently we only have 60 samples in the online database, but I am expecting that to quickly grow as we optimize the lab project. Here is a summary by taxonomic class.
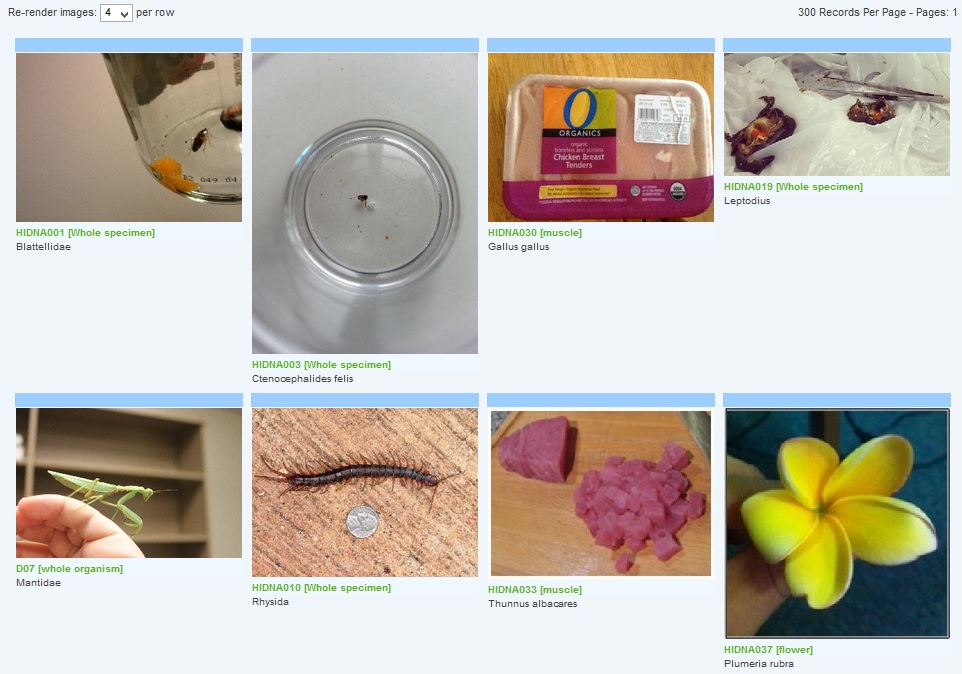
40% of the samples are from flowering plants (Magnoliopsida), while 30% are, not surprisingly, from insects. There is also a project image browser.
And summaries of the data like this accumulation curve.
This shows the slightly diminishing returns as our sample collection progresses. For 60 samples we have (at least) 37 unique species represented (however, several samples could not be identified to the species level). Some species are beginning to be sampled more than once.
I organized the samples taxonomically, again currently only the animal ones, on this page (link) (the page is currently a draft and will need some rounds of cleanup and revision) with links to aid learning more about each taxon and links to the BOLD entry for each sample. I also indicate if the species is native, endemic (only found in Hawai'i), brought here by the ancient Polynesians, threatened or endangered (students are given clear guidelines about what they can and cannot legally collect (no coral, no vertebrates except under special conditions, no threatened or endangered species, etc.) the endangered samples in the database were obtained legally (students are also given common sense safety guidelines for collecting)).
Many of the samples could be identified down to a species level, but some could not, which can raise a flag of interest to check out further as a possible new species. I suspect the mantis sample is a Chinese mantis (Tenodera sinensis) but I am not certain and there are some look alikes among this group so I left it as unknown at the genus and species level. On the other hand, one of the students brought in an insect that seems to fit into the cockroach family (Blattodea) but is not turning up any obvious close genetic relatives on the databases (BOLD and GenBank). A similar story goes for a Xanthid crab and for a hover fly. However, as more samples are collected and the database grows we might get some matches in the future from positively ID'd specimens that will help resolve this.
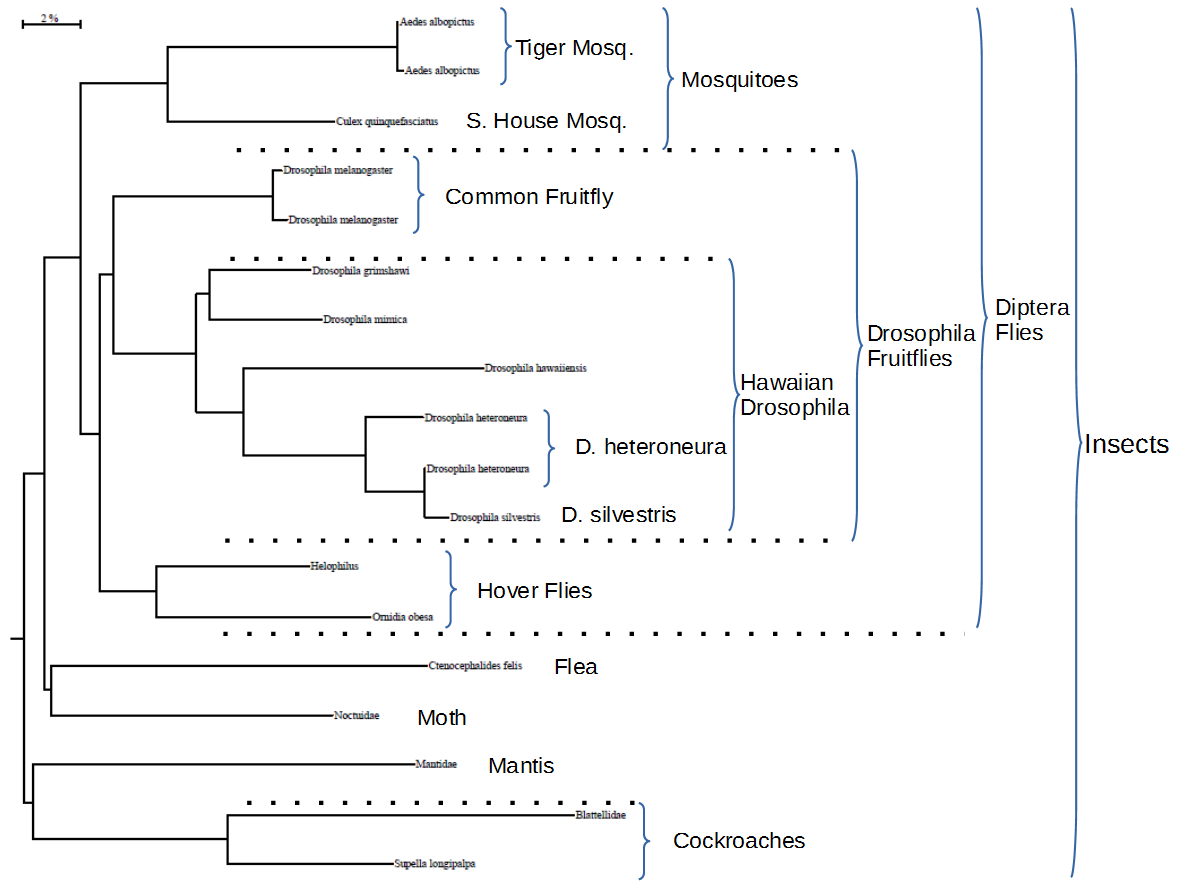
Finally, I couldn't resist generating a phylogenetic tree using our data and BOLD's online tools. DNA barcoding is not designed to behave well in terms of reconstructing more ancient relationships among species. It is aimed at resolving species identifications. So I was surprised at how well the data preformed in clustering the insect species (which contain our largest number of animal samples).
The bar at the upper right represents a distance in the tree of 2% sequence divergence. The tree is rooted with a crustacean (an arthropod but not an insect) sequence (not shown). The flies (Diptera) were grouped together and within this we get mosquito, fruitfly, and hover fly groups. Within the fruitflies we also get a Hawaiian Drosophila clade. Finally we see that one Drosophila heteroneura sample is distant from Drosophila silvestris while another D. heteroneura is quite close to D. silvestris. This is not as surprising as it might first appear because these two species are found to hybridize quite frequently in the wild.
A Hawaiian Octopus and Cowrie
This is a rare find. In another video that I made with my underwater fishing pole camera (see earlier post here), I (unknowingly) went right by an octopus (he'e in Hawaiian, also commonly called by the Japanese name tako here in Hawai'i) in a shallow Mokule'ia reef on O'ahu. He/she blends in well but the suction cups on the upturned arm give it away. This might be a day octopus (Octopus cyanea) but this is really just a guess. Clicking on the image below will link you to the original video.
Even better, the octopus has caught a cowrie! It is hard to see but as the camera passes over you can catch the spotted pattern of (probably) a reticulated cowrie (Mauritia maculifera) just behind and under the octopus. Here in Hawai'i, octopi love to hunt cowries (leho in Hawaiian) and the cowries grow to some of the largest sizes in the world! However, large ones are now rare in the waters around O'ahu. In the image below that octopus is out of focus in the foreground and the mottled pattern of the cowrie shell is visible just behind it. (There is also a sea urchin which gives a size comparison.)
He'e love leho so much that they are used as a lure for catching tako. You can read more about that in the links from here, here and here.