The latest plasmid I tried out for bacterial transformation works well and has a cool end result. The plasmid, pVIB, was constructed with genes from a marine bacteria Aliivibrio fischeri by Engebrecht et al. 1983. This bacteria is bioluminescent and lives in symbiosis with some fish and squid species allowing them to glow in the dark. The enzyme that produces the light is called a luciferase. The plasmid also contains a gene for ampicillin resistance to allow transformed E. coli cells to be selected. So I did a heat shock transformation with pVIB and competent cells, like I blogged about earlier (here and here).
In the image above the upper right and lower left diagonal plates are E. coli spread on luria broth (LB) plates without any selective agent. This is both with (lower left) and without (upper right) a plasmid added to the mix. It might be hard to see but it is just bacteria growth all over, coating the surface. This is referred to as a "lawn" of bacteria. In the upper left an antibiotic (AMP, ampicillin) has been added to the media and there is no growth. The cells have been completely killed. In the lower left the plasmid (pVIB, containing a gene for AMP resistance) has been added and only cells that have taken up the plasmid can grow on the AMP plates.
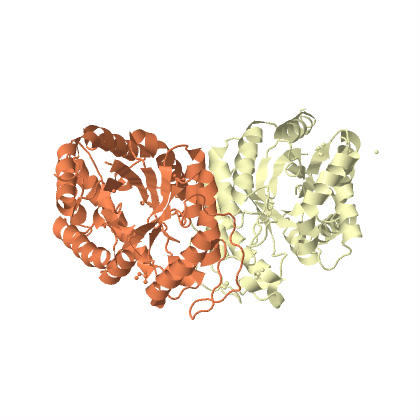
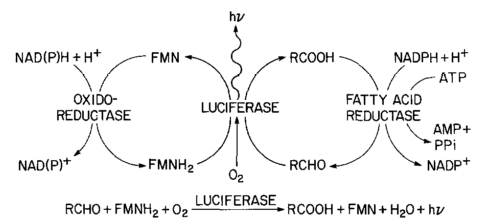
This plasmid contains the genes to produce bacterial luciferase (a dimer (two proteins bind together) with α-luciferase and β-luciferase subunits. The structure was determined by Fisher et al. 1996 (link to structure at protein data bank (PDB) and the PDB molecule of the month). Actually there is a operon (expressed together) of five genes including the two luciferase units. The operon is symbolized by luxCDABE, which gives the order of the genes. luxA and luxB are the alpha and beta subunits. The other genes convert long chain fatty acids into aldehydes with ATP.
From the PDB site you can visualize and rotate the 3D structure of the enzyme.
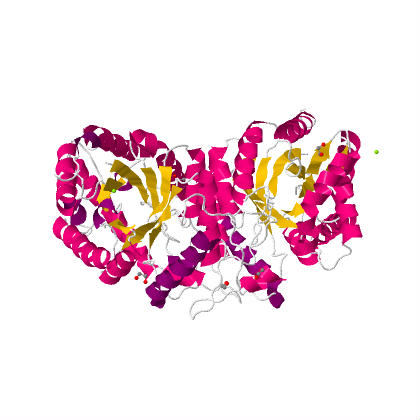
The image above shows the alpha subunit on the left (which contains the active catalytic site) and the beta subunit on the right. These two proteins bind together and create the enzymes quaternary structure. Below is the same view but the components are color coded by secondary structure (alpha helices in red and beta sheets in yellow).
You can see that there is a similarity between the alpha and beta units. It is hypothesized that the beta unit originated as a duplication of the alpha unit and it enhances thermal stability of the enzyme (a fusion protein of the alpha and beta units is sensitive to high temperatures, Escher et al. 1989).
Also, here is a webpage (and here) that contains a great deal of information about bacterial luciferase (there are lots of types of luciferse, like the one in the firefly, but they have different structures, etc.).
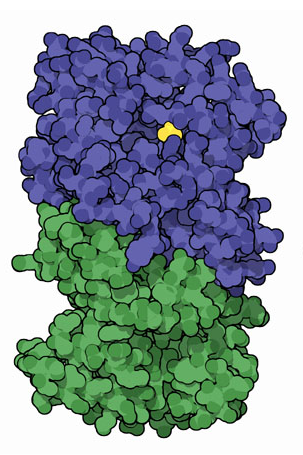
In this image above the active site that catalyzes the reaction is indicated in yellow, within the alpha-subunit (blue).
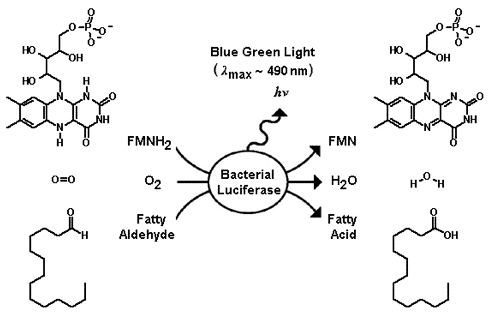
The enzyme takes luciferins, in this case a reduced flavin mononucleotide, and a long chain aldehyde, and oxidizes them with molecular oxygen to produce flavin mononucleotide, water and a fatty acid. There is an excess of energy in the reaction that is released as light with a peak wavelength at around 490 nm (nano-meters) which is a blue-green color.
The additional genes in the operon convert the fatty acid back into an adlehyde. luxC is a reductase, luxD a synthetase, and luxE a transferase; and these assemble together into a fatty acid reductase enzyme complex.
Below is the larger reaction showing the fatty acid being converted back into the aldehyde for the luciferase reaction (and the flavin mononucleotide being reduced to recycle back in).
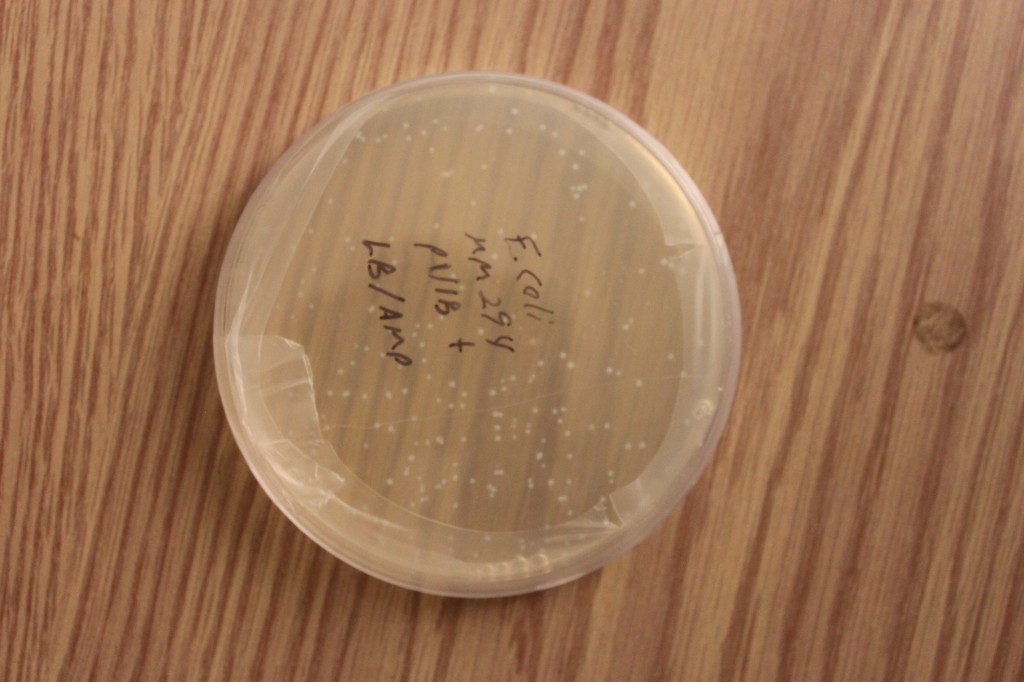
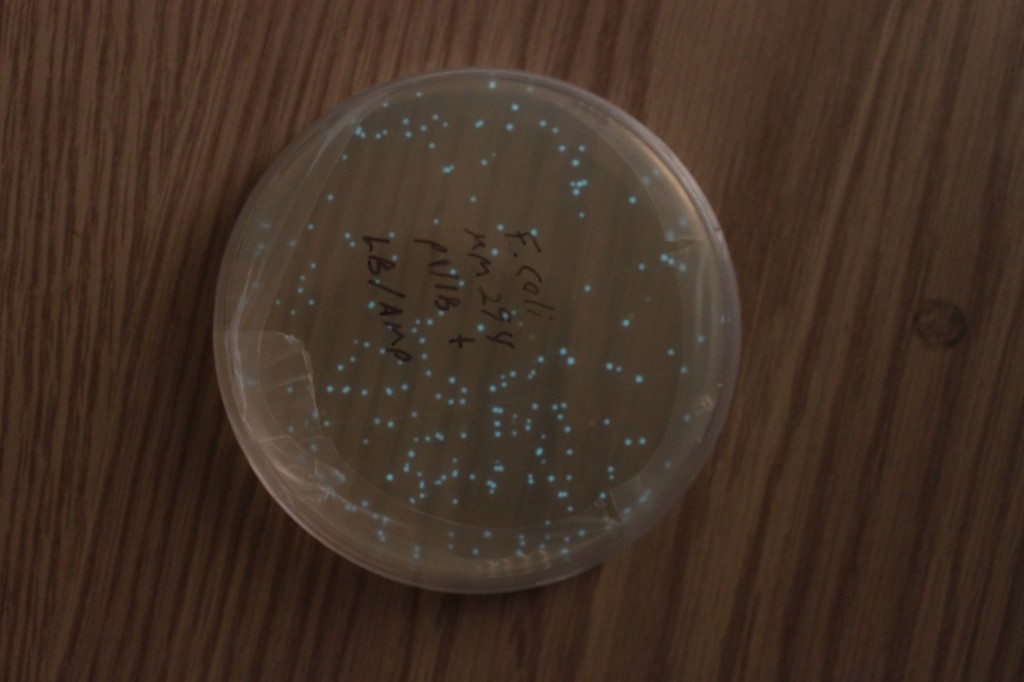
Below are a series of pictures showing the transformed cell clones in progressively lower light levels. They look like normal E. coli cells in bright light.
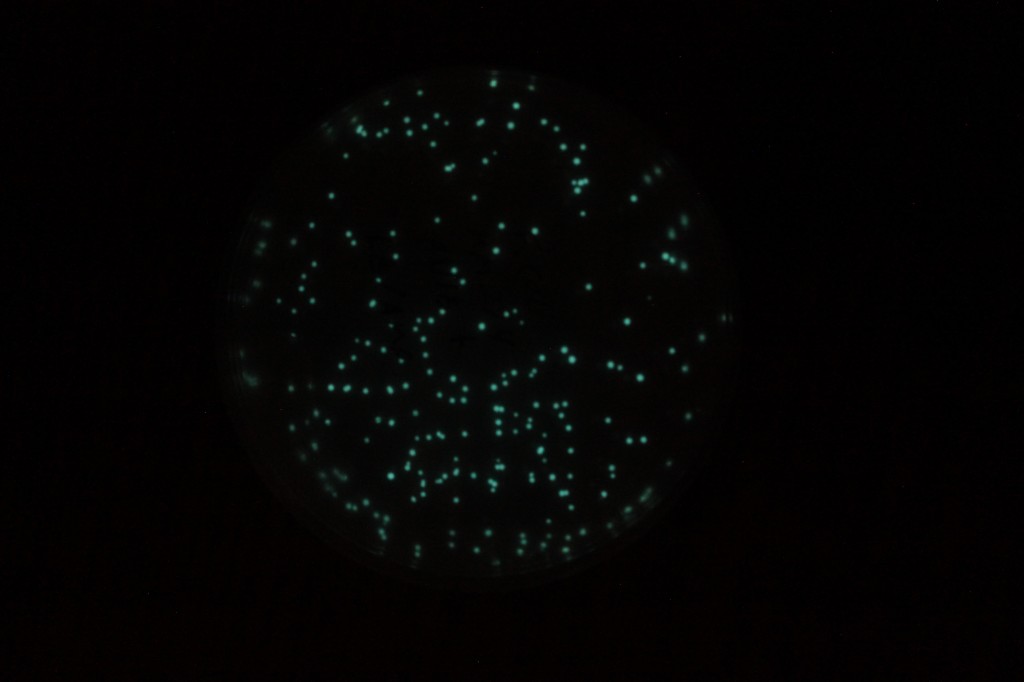
The last picture is an exposure of several seconds. The glow is visible by eye, but you need to be in a dark room and let your eyes adjust for a minute.
The glow doesn't last forever. It was strong 24-48 hours after the transformation. Producing the luciferins requires a lot of the cells energy in the form of ATP for the reductase and the effect is temperature sensitive. I put the plates at 4°C for 24 hours to preserve the bacteria then took them out and the glow was gone. However, I warmed them up to 30°C for an hour and it was back almost as strong as before. I stored them at 4°C again for over a week, then warmed them up again and the glow was completely gone.