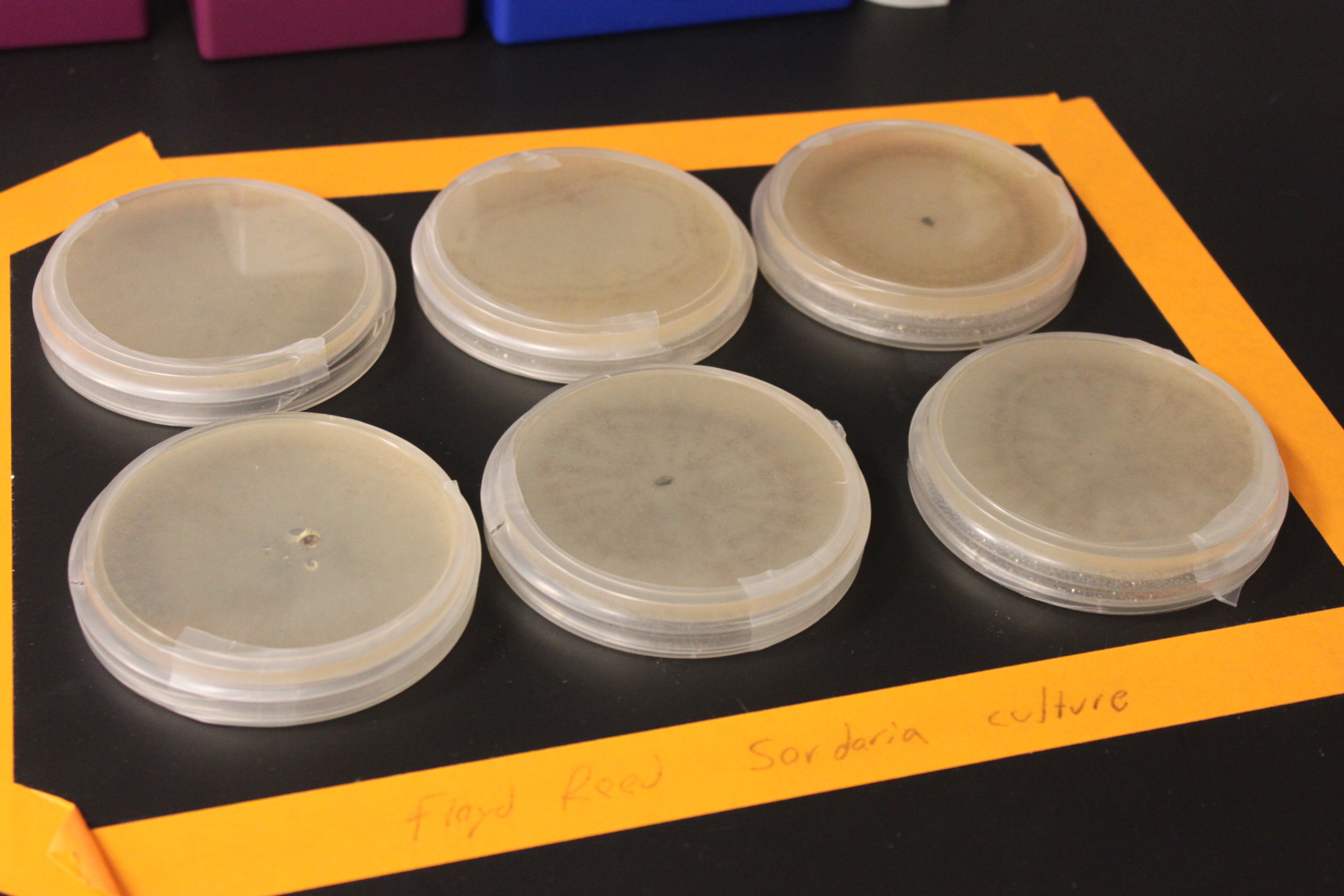
In another lab project I am considering for the fall class, I have been experimenting with crossing Sordaria fimicola fungi. These are molds in the huge phylum of ascomycete fungi that have spores in filaments (asci), which are ordered meiotic products. So it is an excellent example of meiosis and genetic recombination--if you can get it to work.
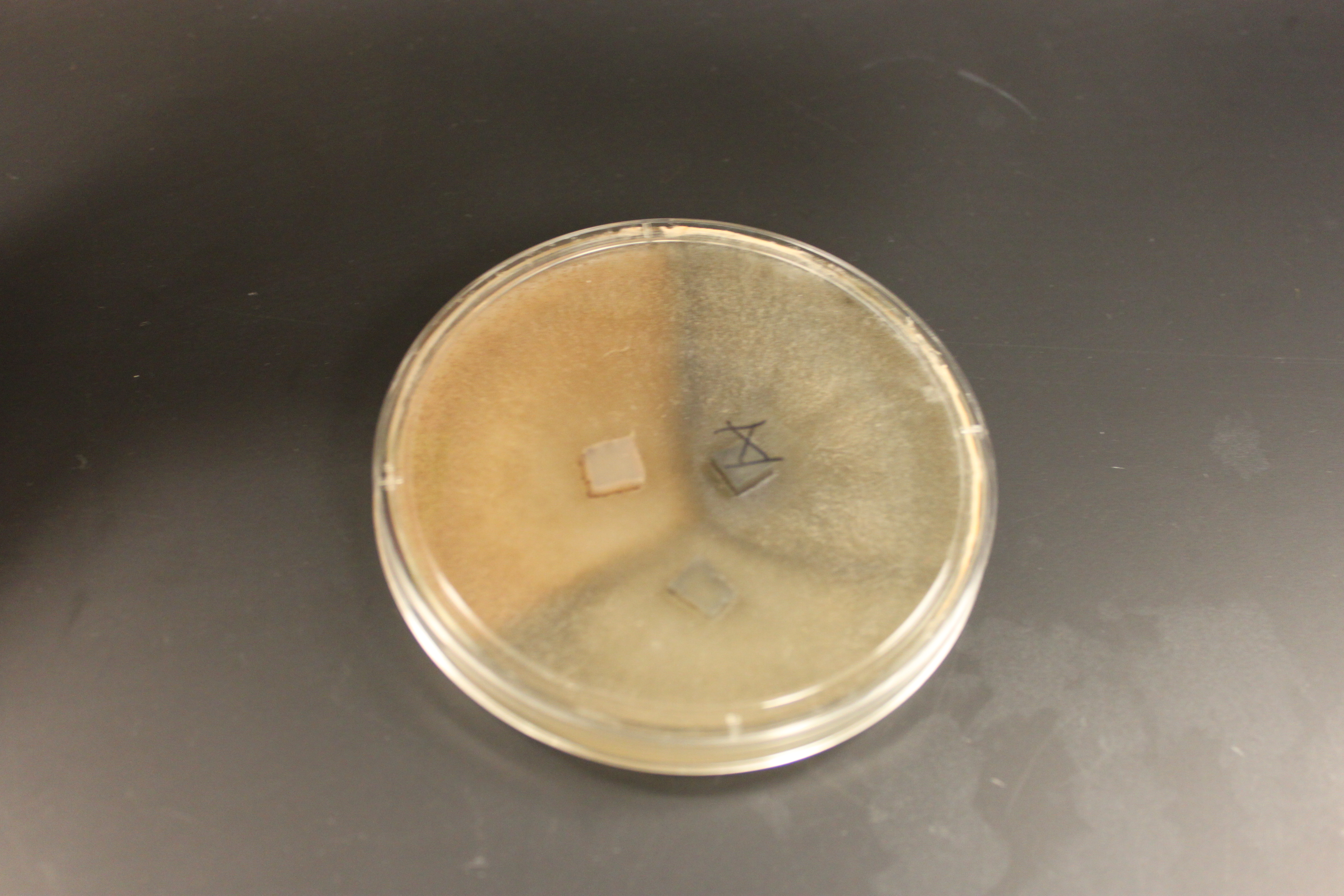
In the picture above I have two plates each of wildtype (lower middle and lower right), tan mutants (upper middle and upper right), and gray mutants (upper and lower left) growing from inoculating the center of each plate with some spores. I had to leave them out on the lab benchtop at room temperature for a few days, so I taped off the area and labeled it in case anyone had any questions about the moldy petri dishes. In the wildtype and tan plates you can see some concentric rings that indicate the temperature fluctuations in the building over the weekend.
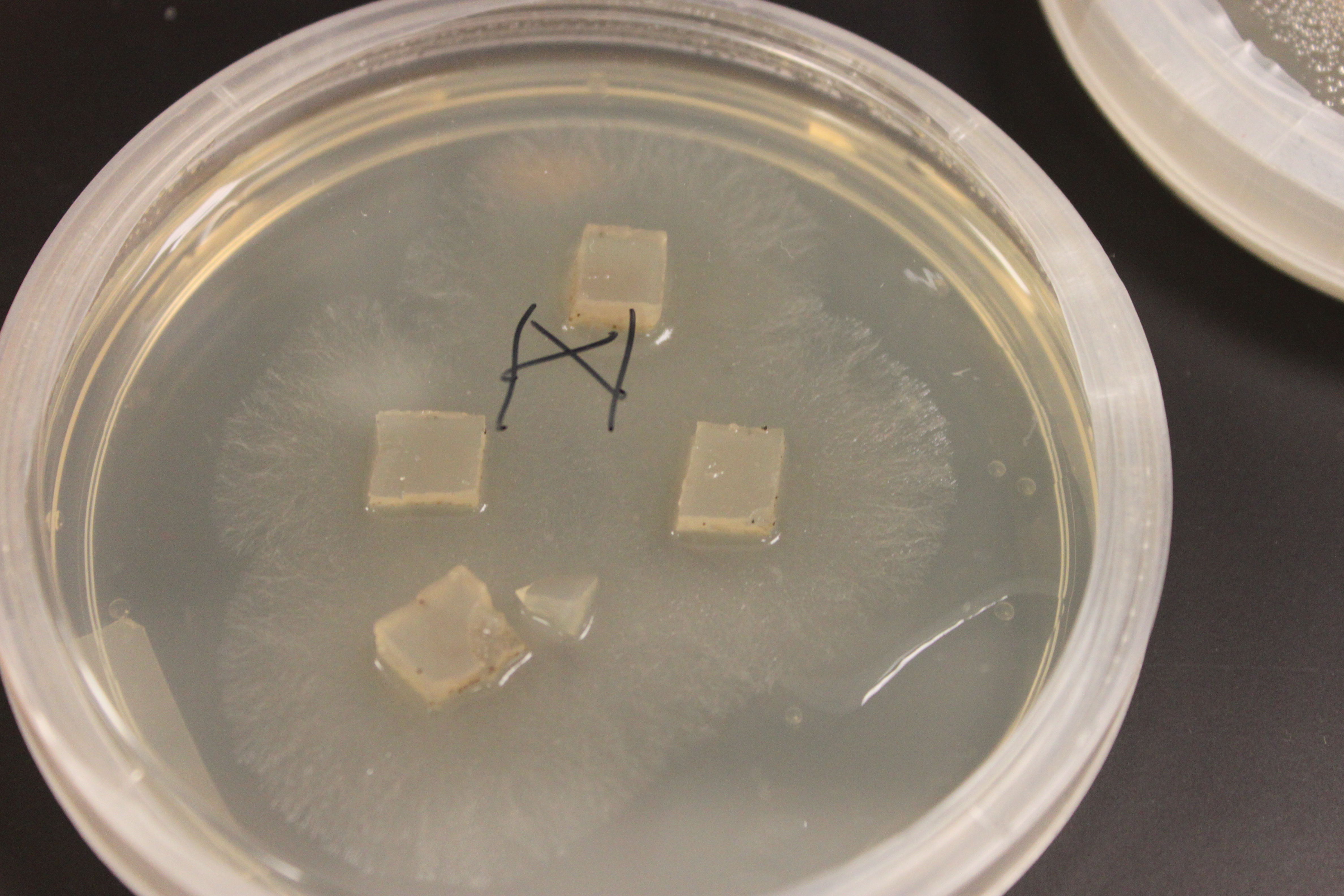
In the picture above and below I have set up crossing plates by cutting out cubes of agar containing growing fungi of different types and placing them upside down on the new medium. You can see the mycelium growing out in a circle from each cube.
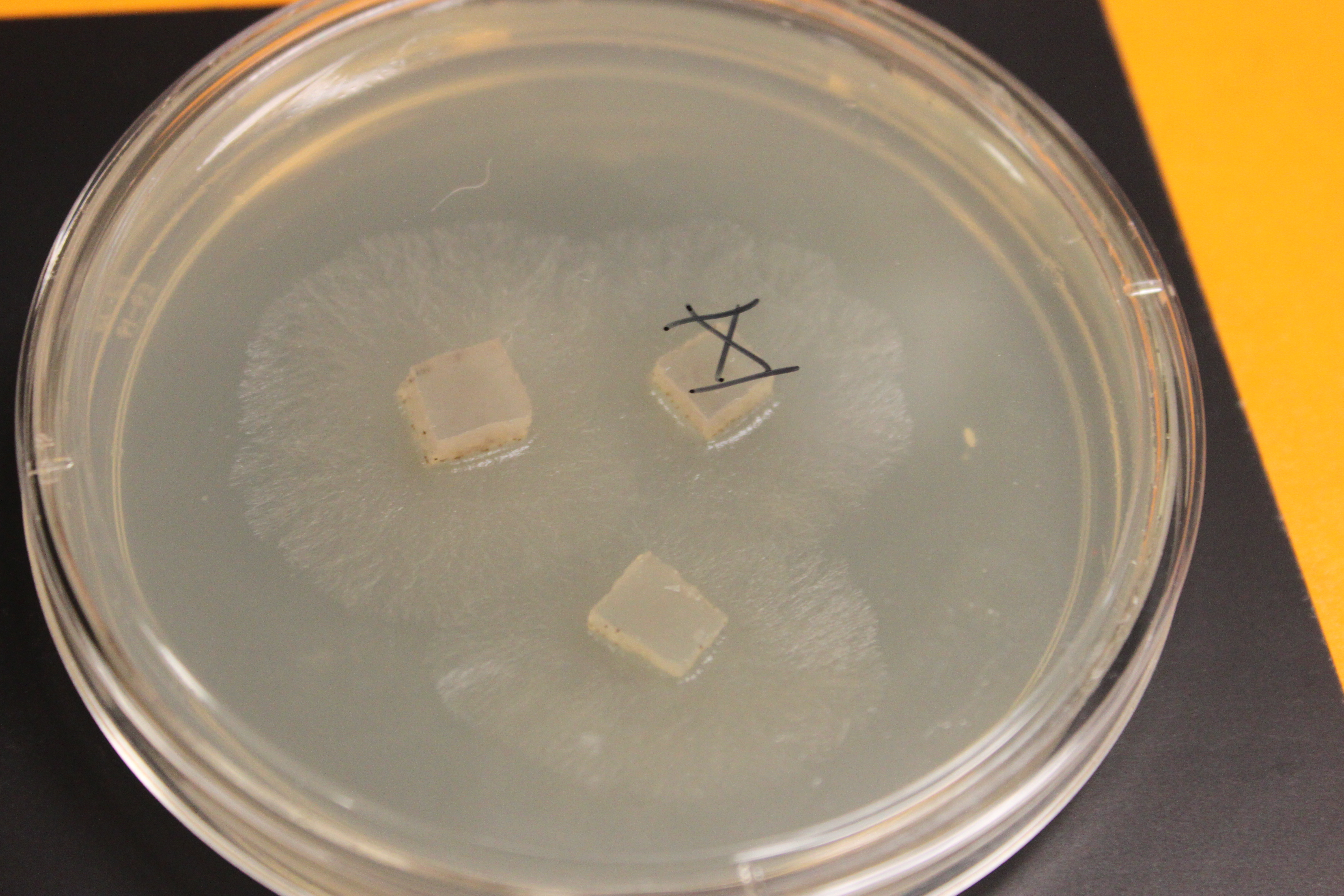
Below are older tan and gray mutants that have grown into each other and are crossing. The darker X at the border looks like wildtype and might possibly be an example of genetic complementation but it is hard to tell if this is not also just denser growth.
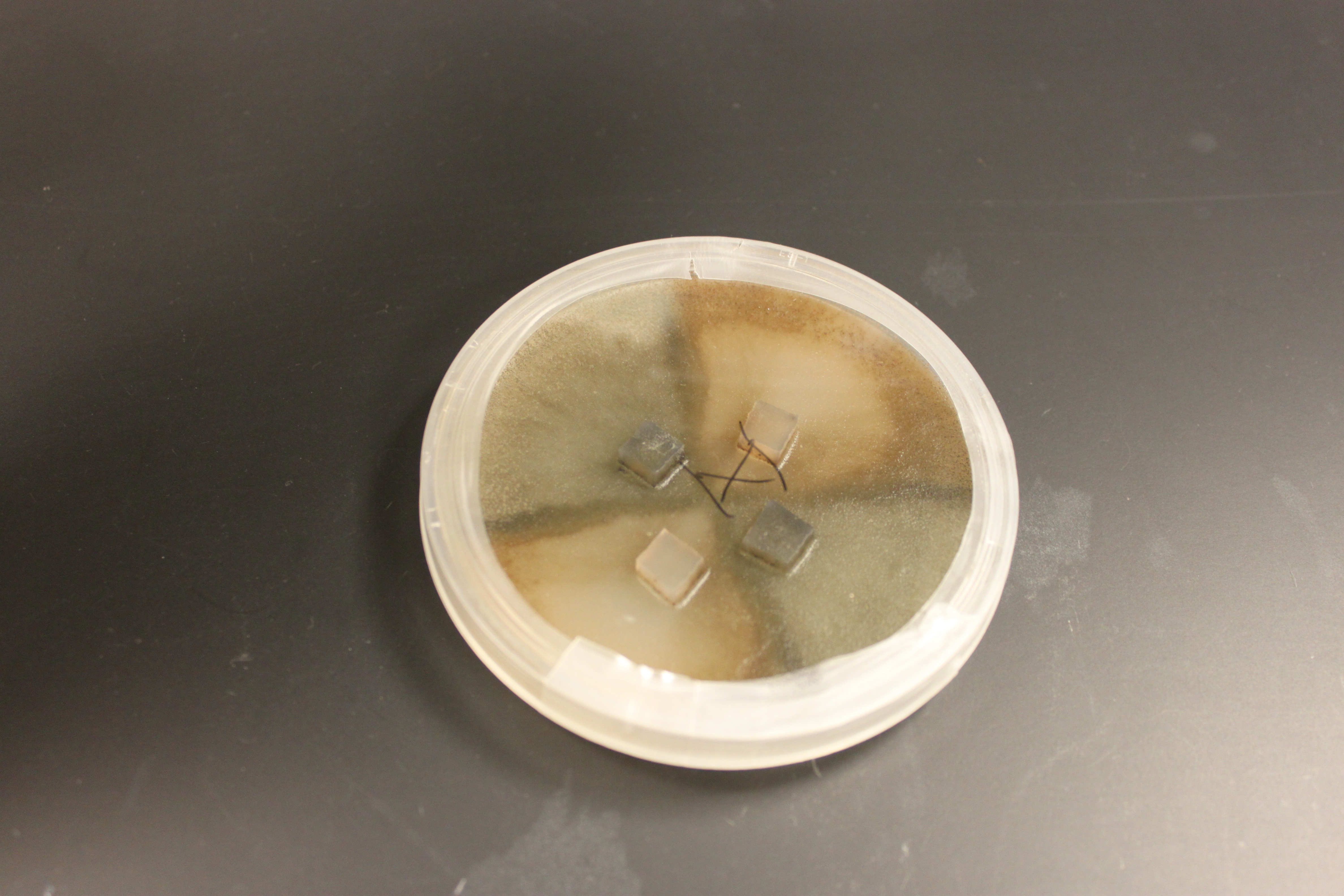
And in the plate below is a cross of all three types. Wildtype in the upper right; tan in the upper left; and gray at the bottom. (Note that the boundaries with wildtype are darker than wildtype alone, which suggests denser growth is at least partially responsible.)
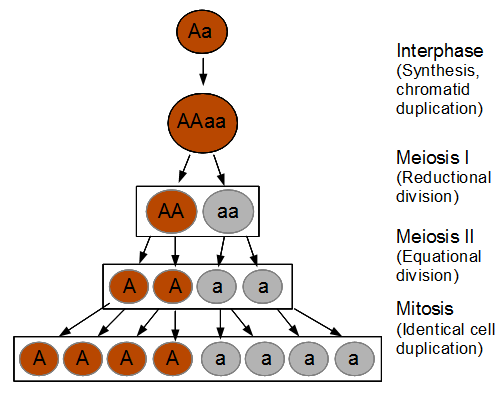
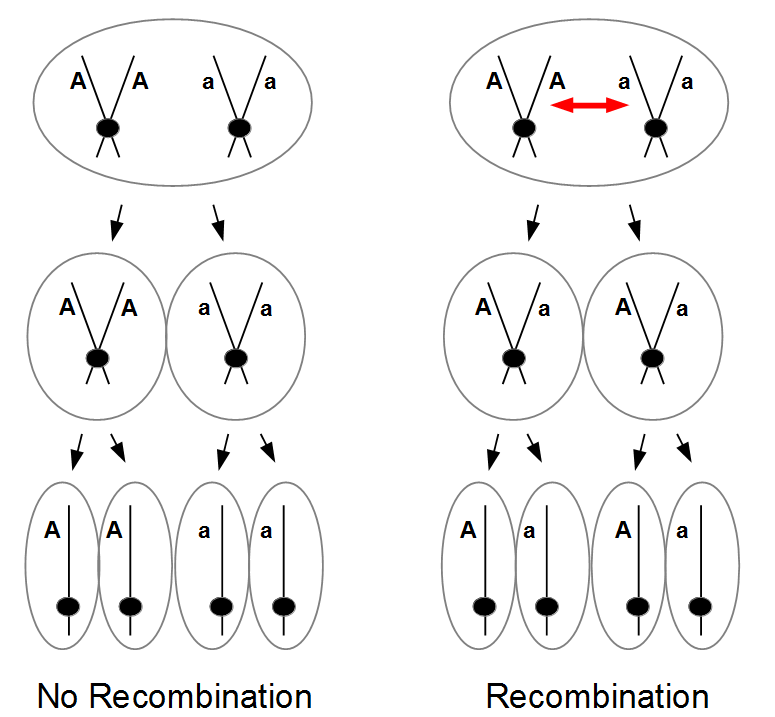
The spores are interesting in this group of fungi because the meiotic products remain oriented relative to each other according to the pattern of chromosome segregation. The the effects of recombination between the chromosome's centromere and the gene causing the difference in spore color is directly observable. To illustrate I've diagrammed meiosis in a heterozygote below (these fungi also have a round of cell duplication by mitosis at the end of meiosis resulting in eight spores from each starting diploid cell).
So the reductional division in a heterozygote leads to a 22221111 (or 11112222) pattern of ascospores. In meiosis I homologous chromosomes segregated (followed by sister chromosome separation in meiosis II) which leads to the four and four ordered pattern.
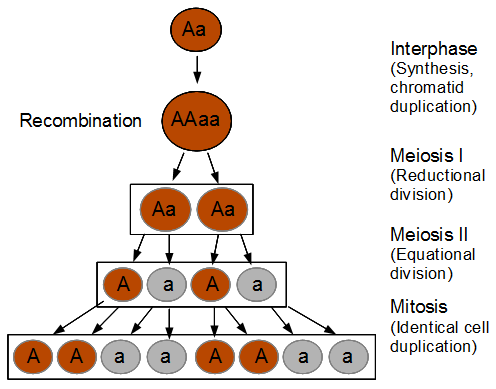
Below lets look at what happens if there is a recombination event.
Recombination exchanged parts of the homologous chromosomes, so the duplicated alleles were moved to different chromosomes to segregate away from each other. So in the end the four-and-four pattern is broken up.
In the figure I have shown a 11221122 pattern but this could equivalently have been a 22112211, 22111122, or 11222211 pattern as well as a result of recombination. The key is that both spore types appear in each half of the asci. Why does this happen with recombination? The chromosome segregation (and chromatid separation) are controlled from the centromeres (microfilament fibers attach to the centromeres and they move apart to opposite side of the dividing cell).
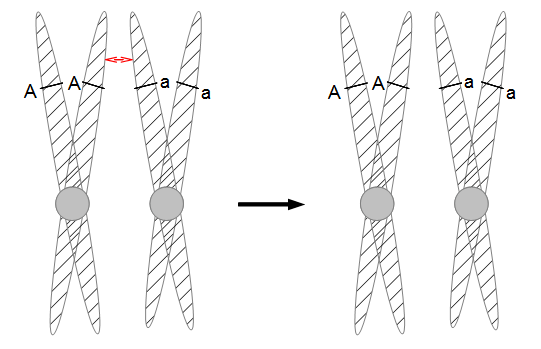
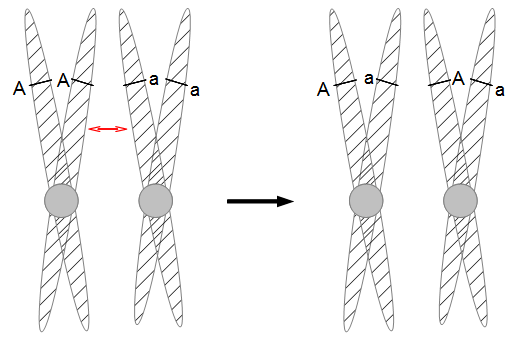
In the figures above and below I have indicated condensed duplicated chromosomes joined at the centromere (circle). The gene's position with the two alleles we can observe is indicated with the line and the alleles with an "A" or "a". Recombination is indicated by a red arrow. Distal recombination beyond the gene, away from the centromere, has no effect on what we can observe (above). However, proximal recombination between the gene and the centromere exchanges the alleles (below).
So as meiosis progresses, the alleles have switched to different (homologous) chromosomes and end up in a different pattern due to recombination. To try to connect the four different figures above I have drawn it a different way below.
I left out the final duplication of each cell into the eight spores at the end. (And this is very stylized, cells and chromosomes don't really look anything like this; I'm just trying to get the idea across visually.)
So the frequency of recombinant meiotic products from heterozygotes gives you an idea of how far from the centromere the gene is located on the chromosome. Normally we count the fraction of recombinants and divide by the total to get the recombinant fraction as a measure of distance. (This also undergoes a long distance correction for multiple recombination events, but I will talk about that later.) However, the Sordaria recombinant pattern spores can be a little misleading. What we are really seeing, usually, is one recombination event out of two possible. We don't count the non-recombinant spore pattern that is present with the recombinant one in the same asci filament. So there is a correction-that is easy to forget-where we divide the fraction of recombinant patterned asci by two.
So that is the theory; how about in practice?
The mycelium growing from a spore is composed of masses of thread like hyphae. These secrete enzymes and absorb nutrients from the environment as they grow. Essentially the mass, which can sometimes become huge in nature, are all considered a single organism (so when I cut out some agar containing hyphae to set up the cross I essentially cut off pieces of a single fungus to regrow again). Like mushrooms and many other fungi the mycelium is often hidden in the soil or whatever material the fungus is growing in. When the hyphae from two different organisms, but within the same species, grow into each other they cross, recombine, and release spores from fruiting bodies like the above ground mushrooms we are familiar with. In Sordaria the acsi form inside tiny round perithecia (the fruiting bodies) that you smash open with a coverslip in a wet mount on a glass microscope slide. If you press too hard the asci shear apart; not hard enough and the perithecia are not ruptured. In addition to this they have to be the right age. Too young and the perithecia do not rupture easily and the ascospores (spores in the ascus) do not have enough pigment to be able to visualize the genotype. Too old and the peritheca spontaneously rupture and eject the asci (even before you get to them--this is what they do in nature but in the lab they stick to the inside lid of the petri dish). When I first tried to look at them they were too young. Then when I aged them a bit and tried again they were too old and were beginning to coat the lids with spores. Here are some imperfect pictures from teaching myself how to do this.
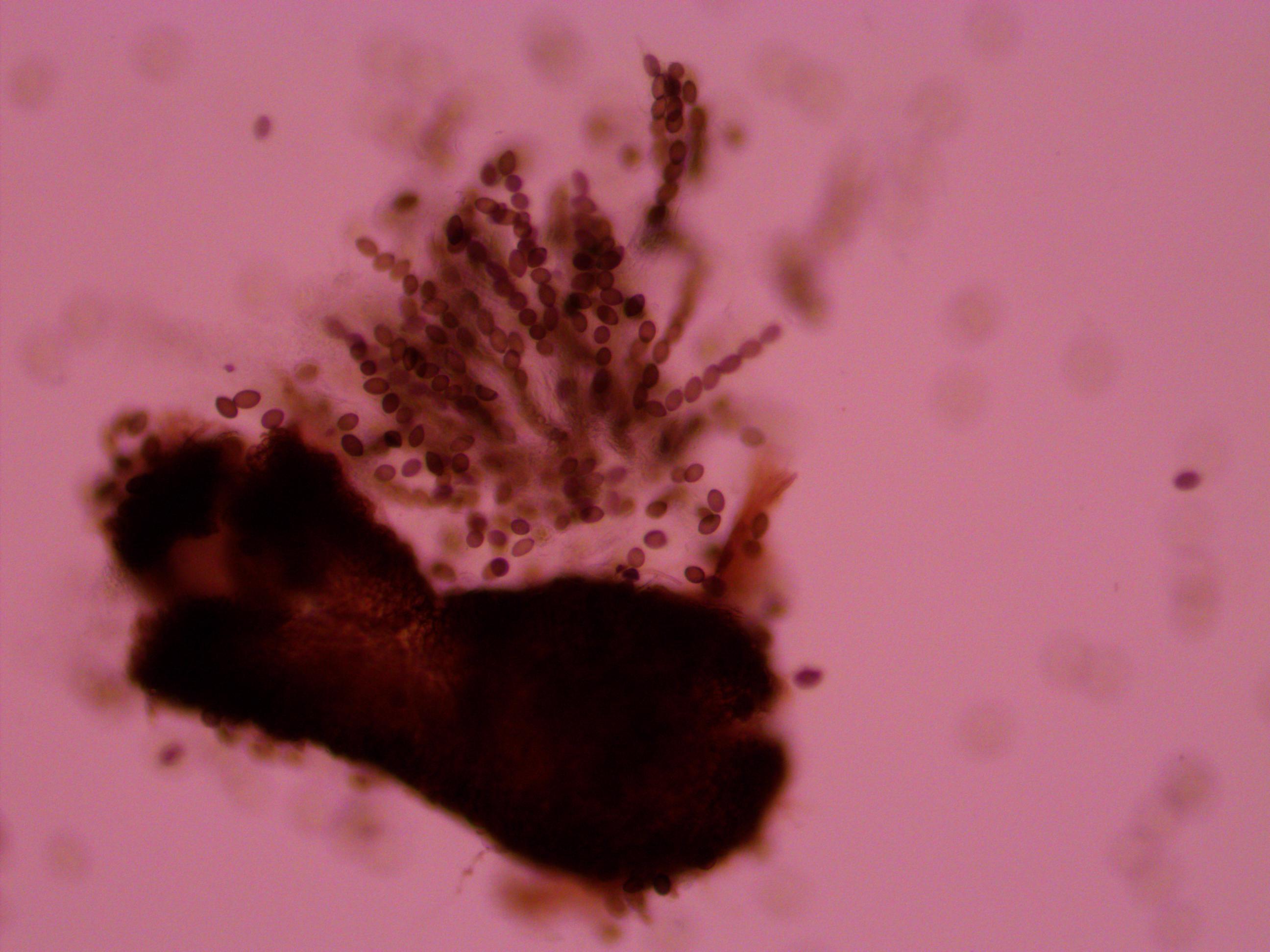
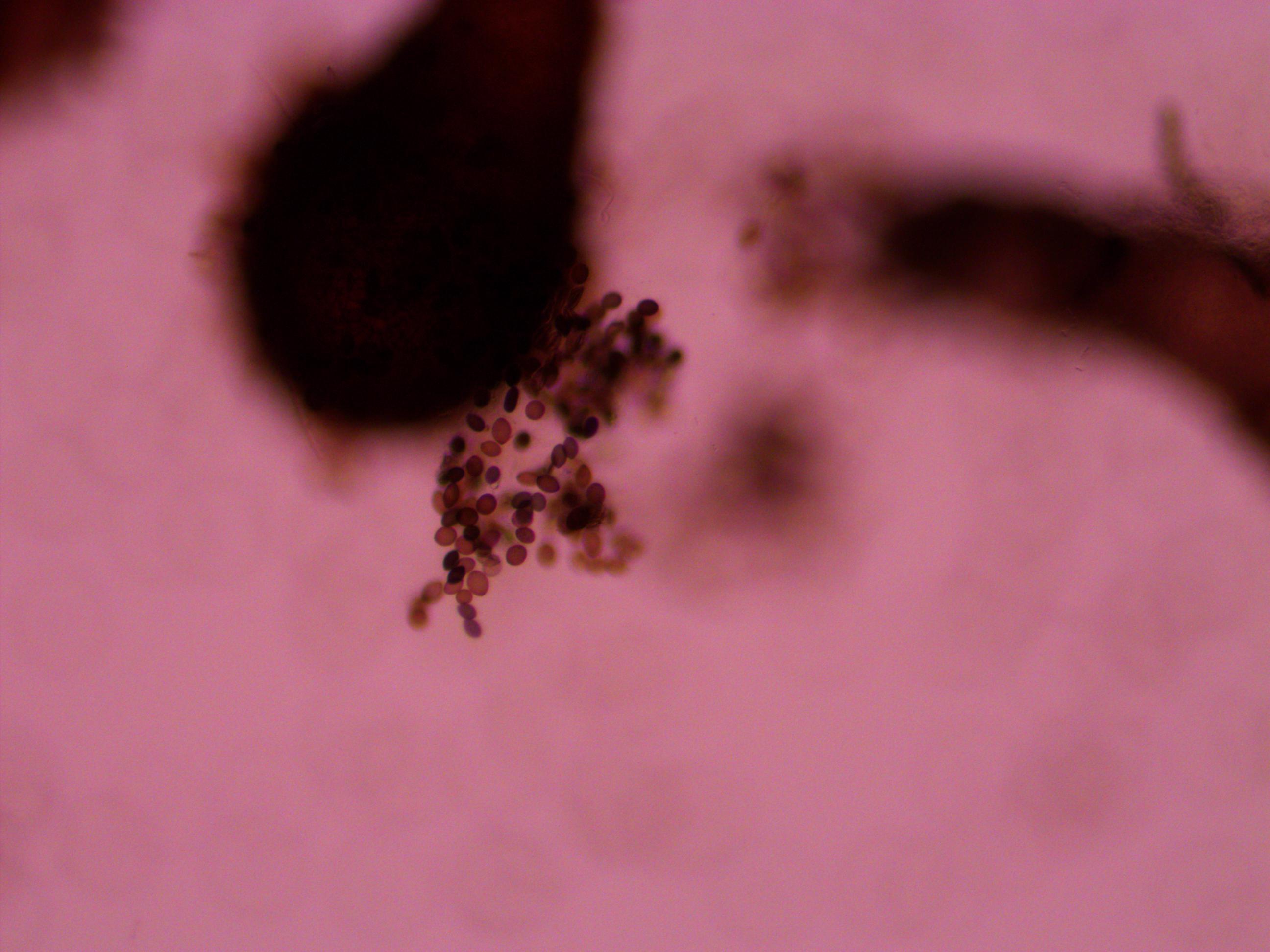
Above is a squashed perithicia with a cluster of tan asci beside it. Below are some darker wildtype spores.
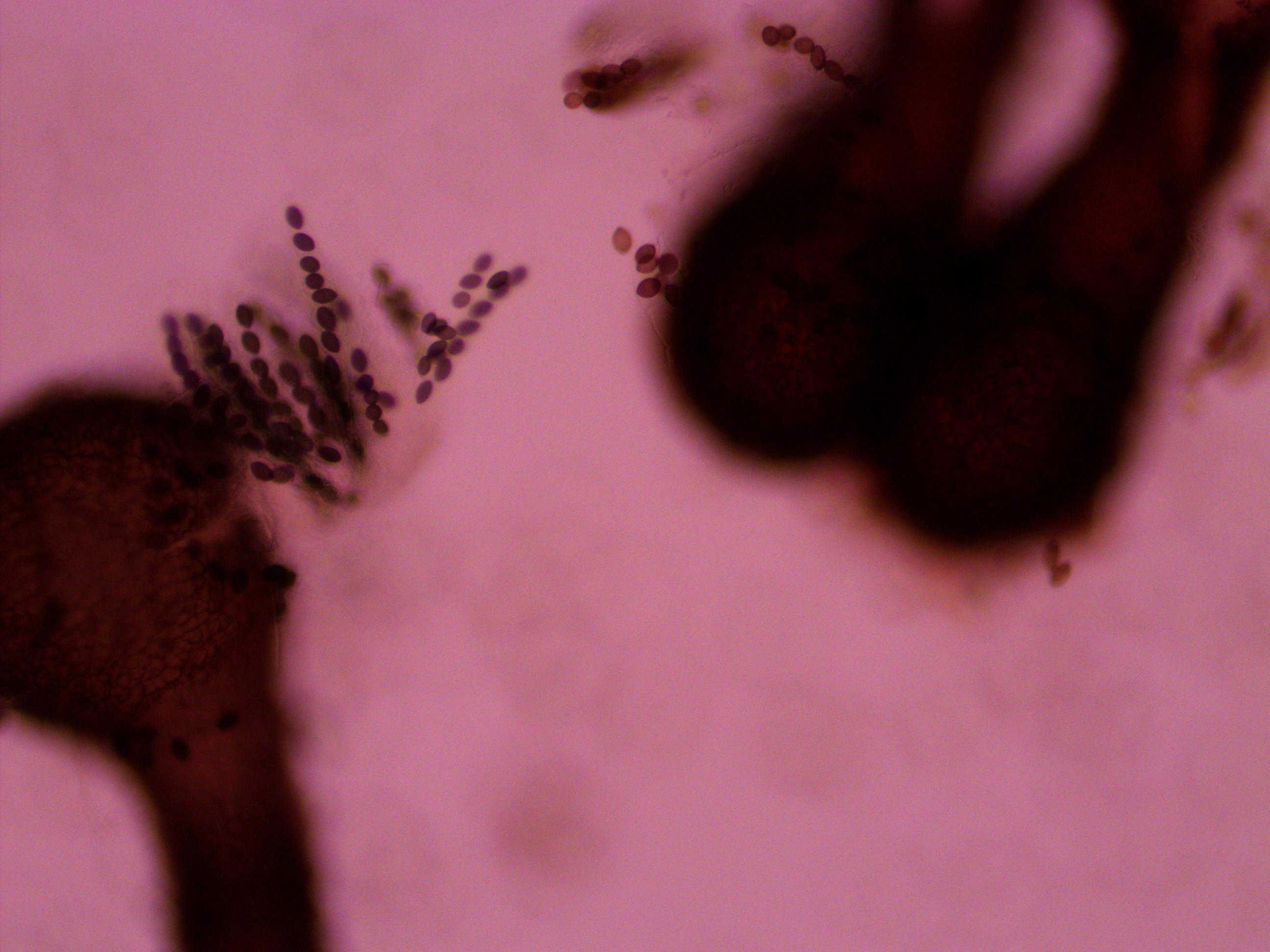
Above you can see both tan mutants and wildtype spore colors, from different perithicia, in the same picture. Below is a mix of alleles but frustratingly I can't tell how they are ordered or if they are just mixed on the slide.
Below is an example of a bunch of loose spores, which happened all too often.
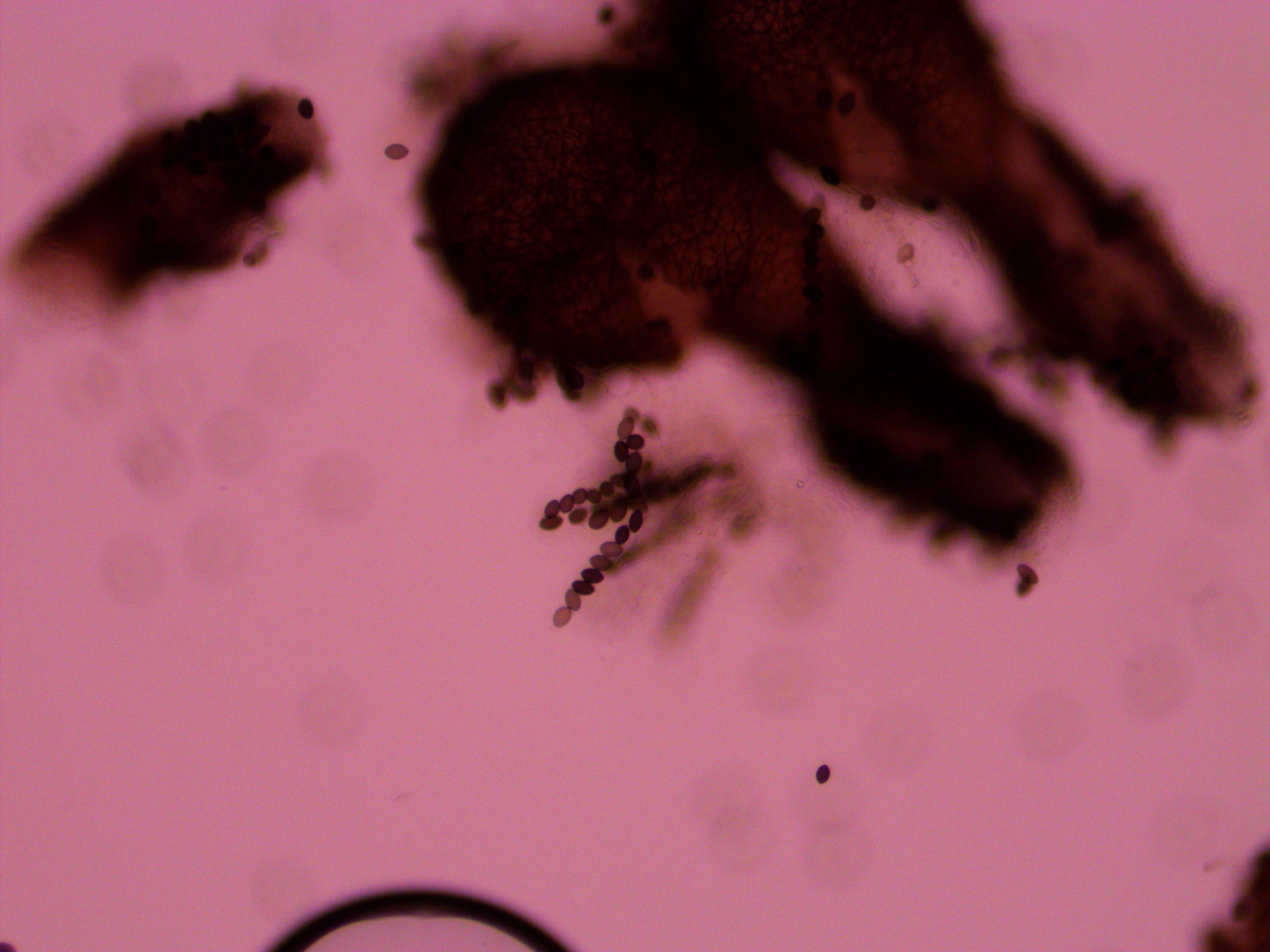
And finally bingo!
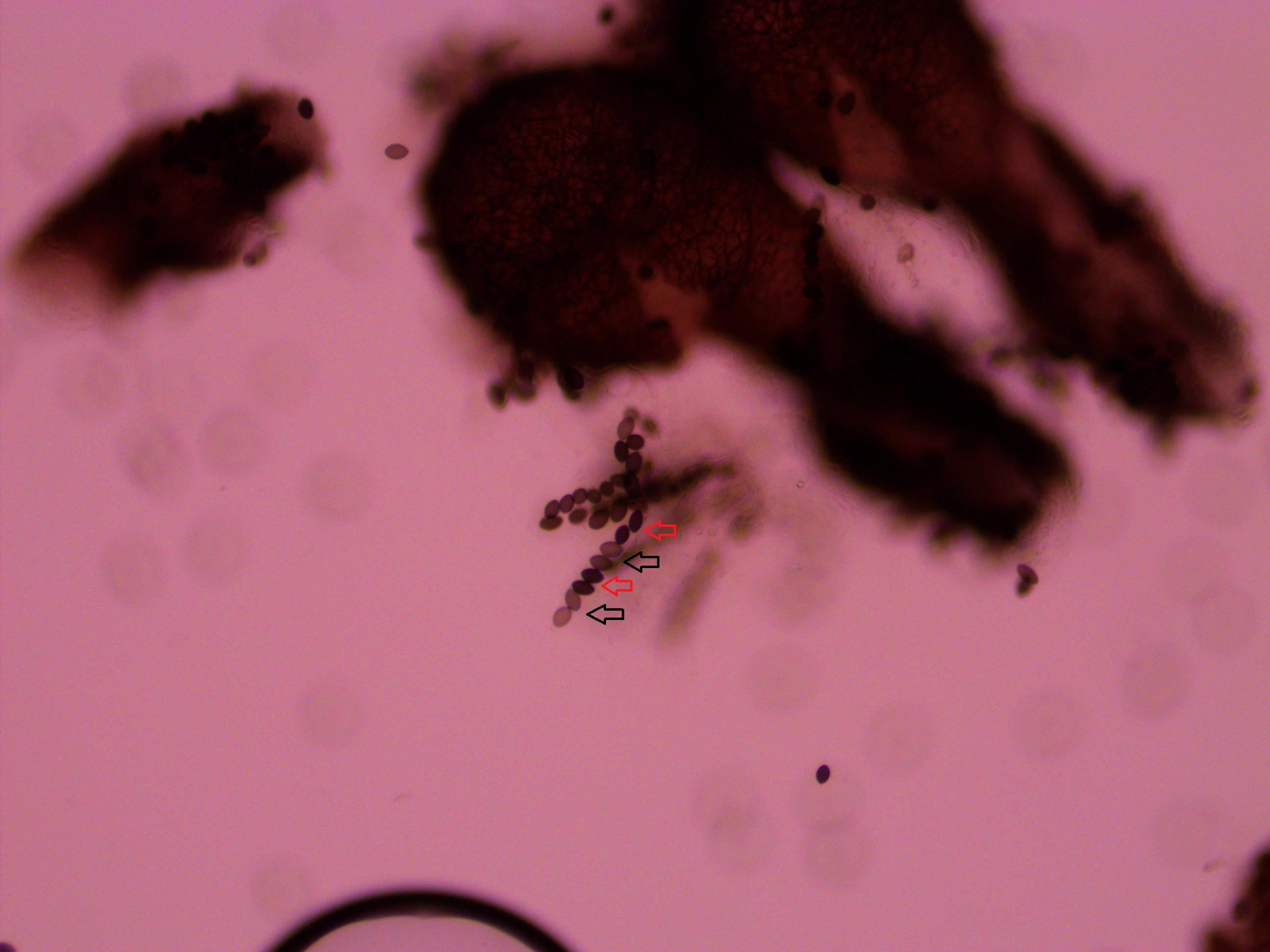
Above is a recombinant meiotic product. The asci has a 2-2-2-2 pattern of wildtype and gray mutant spore colors. I've indicated them with arrows below.
The fourth one down appear a bit darker but that is because of overlap with another asci behind it. I need to keep practicing to get the timing and method down so I can get more useful results with nice flat squashed asci that are not sheared apart. It was nice to spot a recombinant but I can not yet score enough asci to get data for calculating distance from the centeromere.