One undergraduate in the lab, Kenton Asao, is interested in chromosome evolution and has been troubleshooting protocols to prepare insect polytene chromosomes for imaging. I worked with polytene chromosomes many years ago in grad school---but not since---so I am quite rusty and need to brush up on this myself, and this is something we would like to be able to routinely do in the lab. Long story short, it wasn't working starting out so we sent some photos (to make sure we were dissecting out the right tissues) and asked for some advice from Kevin Cook (D. of Biology, Indiana U.) who kindly sent us his protocol. We continued troubleshooting and we were getting quite frustrated trying different stains and imaging approaches but nothing seemed to be working (we also asked for advice from Gert de Couet in the department here at U.H. who loaned us some different dye and reagents to try out). In the meantime we tried extracting DNA and doing some PCR for some damsel flies and that didn't even work...! Then this morning Kenton found me and said that he needed to show me something. After taking a look I went back to my office to get our microscope camera and set it up to record some images.
Some background. Generally chromosomes are very tiny and difficult to image with standard equipment. However, some cells undergo endoreduplication where the chromosomes divide but the cells do not. In some of our muscle and liver cells there are actually four (tetraploid) or eight (octoploid) copies of each chromosome instead of the normal two copies (diploid--one from each parent) that we are used to thinking about. In older fly larvae that are about to pupate and metamorphose into adults the salivary gland chromosomes divide approximately nine times resulting in 512 copies of each of two starting chromosome copies or about 1,024 total copies per cell. In addition they stay physically associated with each other so that they form large (gigantic on a cellular level) structures. Each individual chromosome is made up of about half DNA (actually a single very long strand of DNA) and half proteins that organize the DNA into the structure of the chromsome. The density and composition of proteins changes along the chromosome so that different regions have different staining levels that result in a banding pattern. In polytene chromosomes all 1,000+ copies are organized together into these large structures.
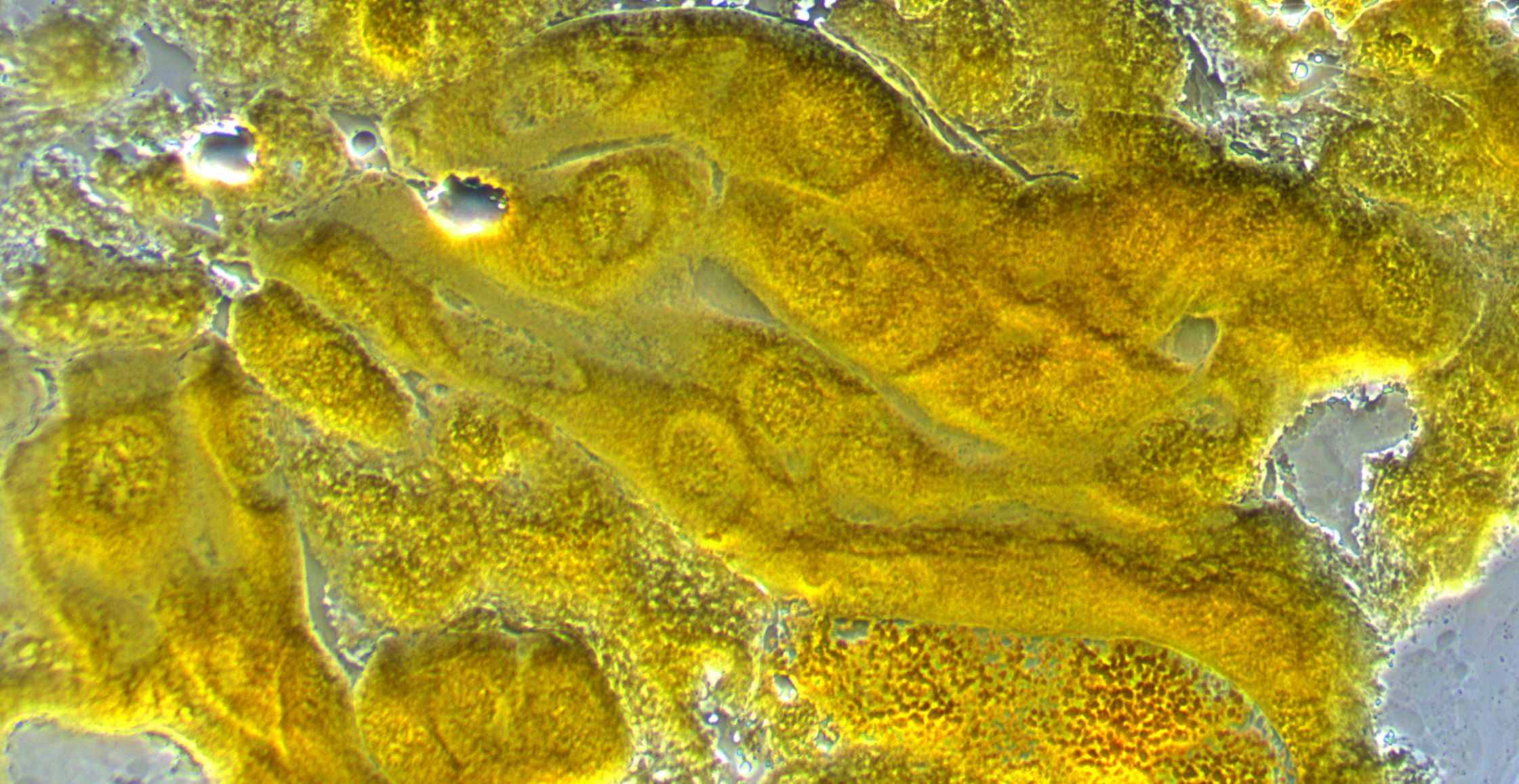
This first image below is of some cells in larval salivary glands under the microscope. Within the cells you can make out the nuclei and maybe the first hints of the darker chromosome structure.
The next image is with higher magnification. You can see the cell nucleus as spheres with darker chromosomes winding around. In preparing the tissue for these images we apply a lot of pressure to purposely try to rupture the cells and spread the chromosomes (i.e., this is not necessarily what they would normally look like).
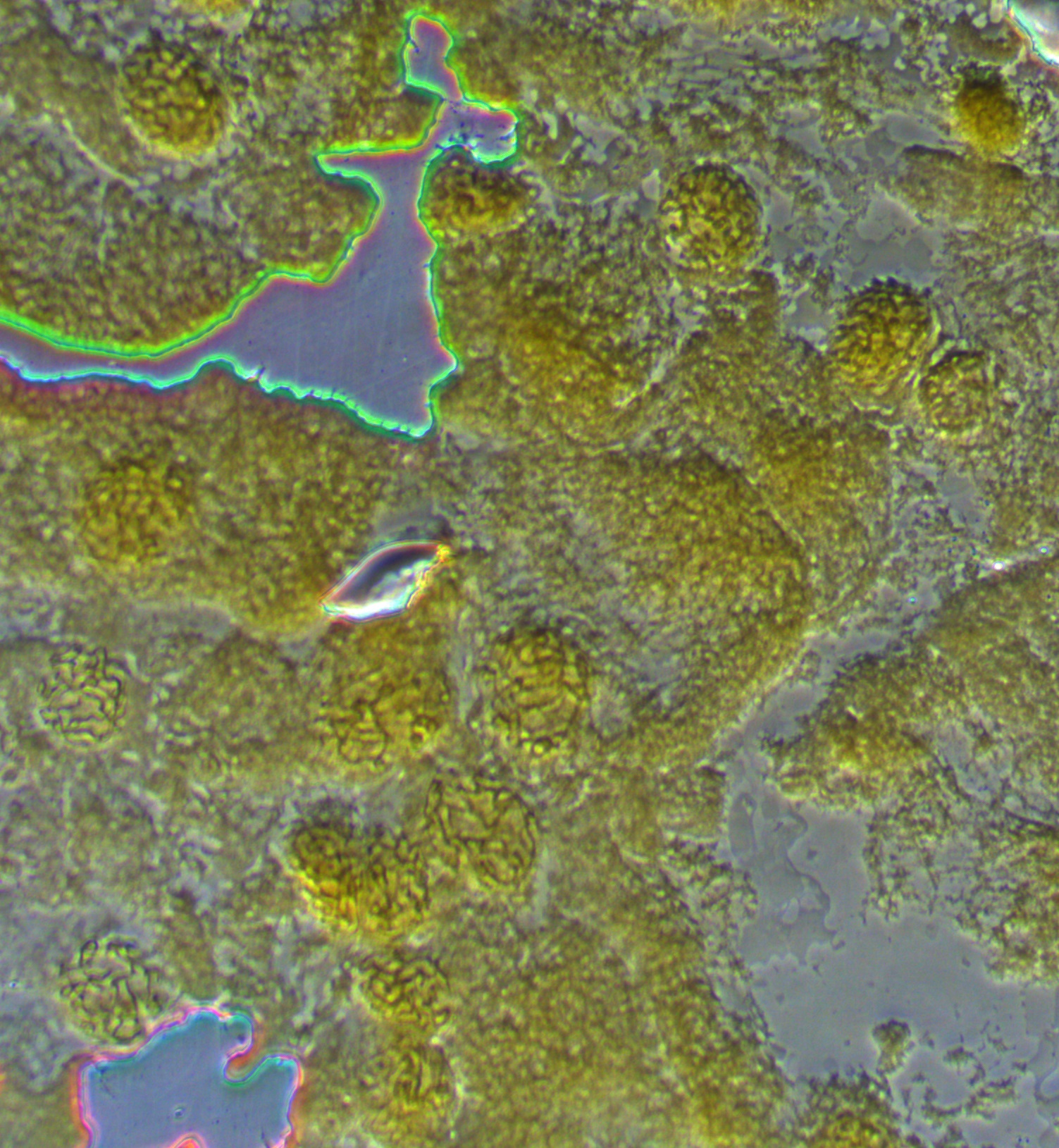
In the image below one of the nuclei has been squashed and "spread" a bit more than the others. The chromosomes look like darker squiggles.
The image below is zoomed in to an even higher level. You can start to make out the dark and light banding pattern along the chromosomes.
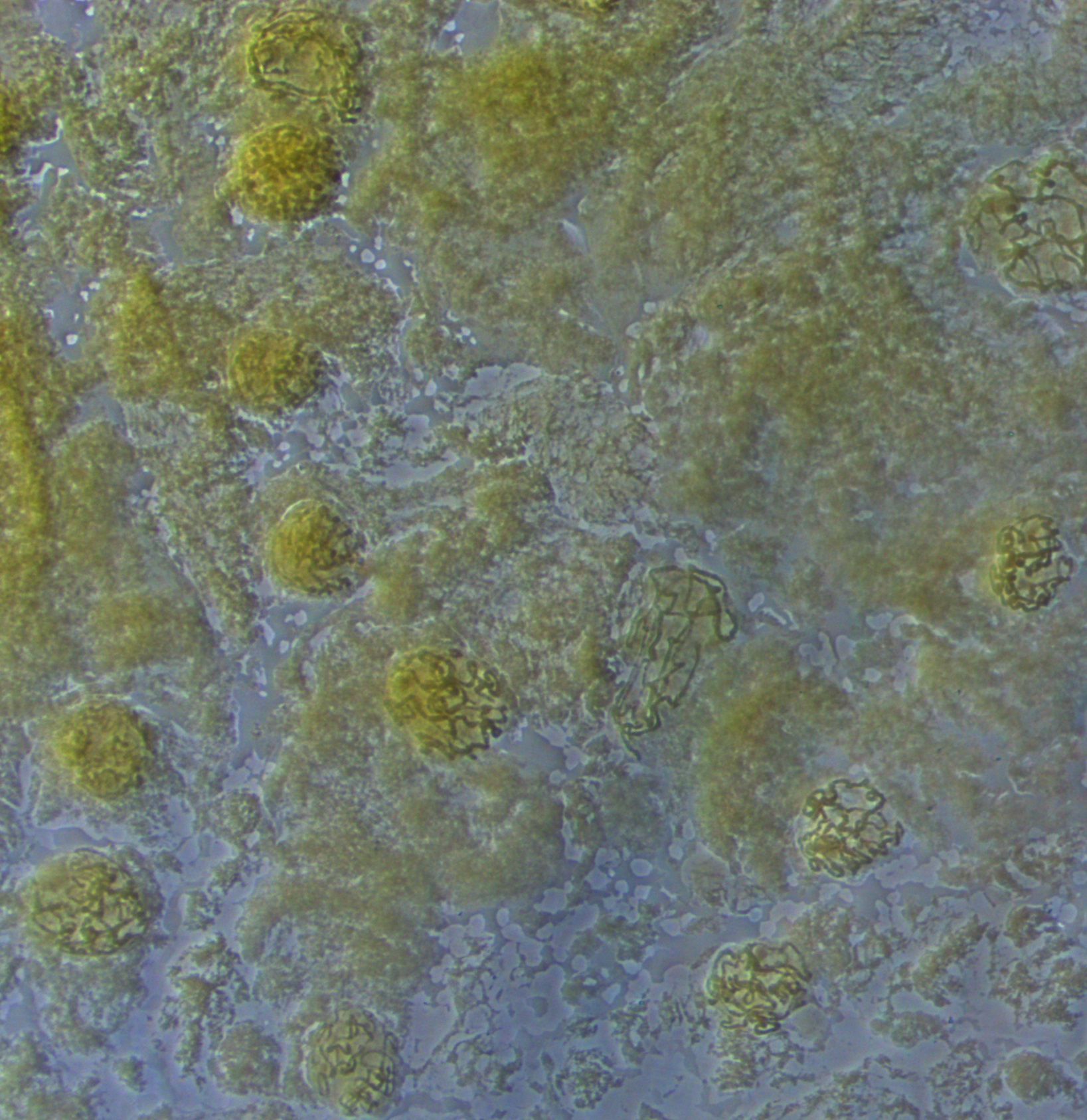
Below are two nuclei. The one on the left has been spread more than the one on the right, and the banding pattern is easier to make out---at least for the section that is spread.
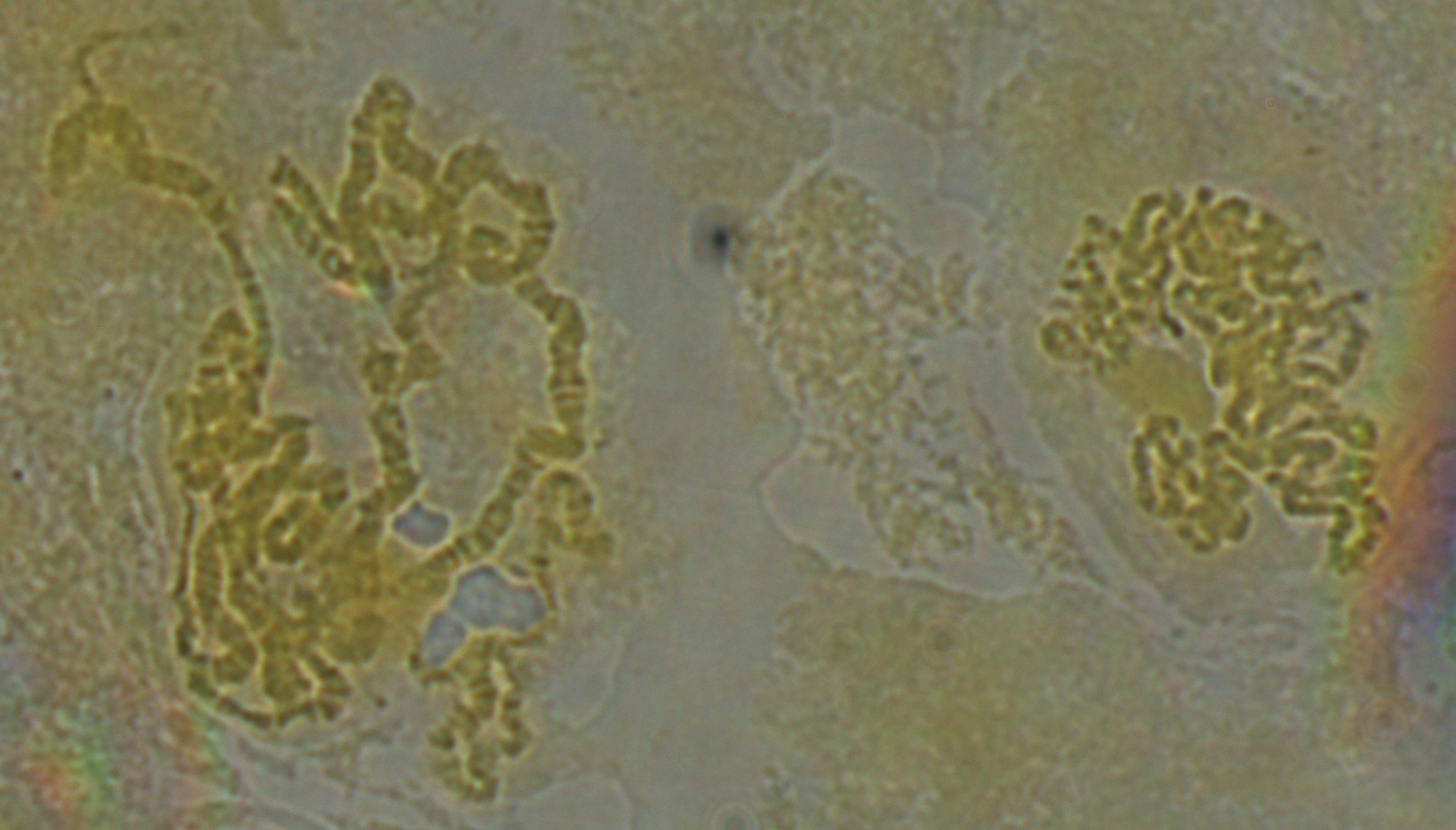
In the image below it looks like some of the strands are starting to shear. However, the end of one chromosome is visible to the right so I wanted to focus on it and see if I could identify which chromosome it belonged to.
Below is zoomed in some more on this area, and this is pushing what the optics in our system are capable of. Just down from the end of the chromosome is a "puff" where it widens out. These correspond to areas with active transcription of genes.
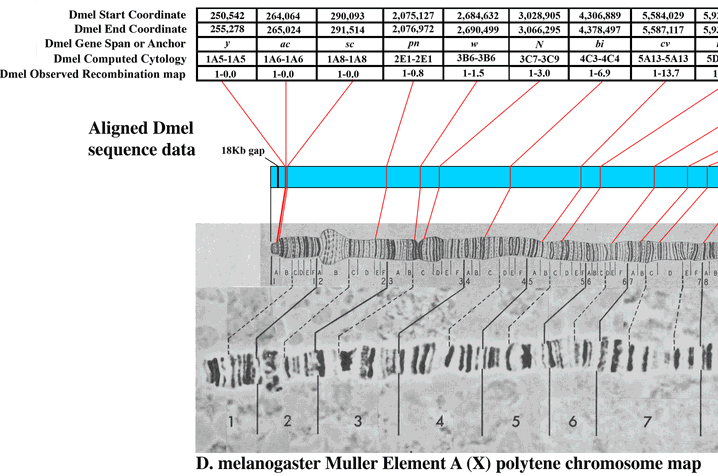
There are cytological maps of the chromosomes available at flybase to compare to. Here is the image at the end of the X-chromosome.
Based on the similarity in the banding pattern and the presence of the puff I suspect this is the end of the X-chromosome. The gene yellow (y) which affects body pigment is at the very end and white (w) which results in red eyes when functioning correctly (and happens to be the first genetic variation discovered in Drosophila melanogaster by T. H. Morgan's lab) is just down from the puff at the second dark band.
Ironically the best images today were not from the orcein stained slides but from Hoechst 33258 stain, but strangely the chromosomes did not appear to fluoresce under UV light. Kenton wrote down all the steps he took today and the most important thing for now is to replicate the process tomorrow to make sure we can get good images of the chromosomes. Then we will try to fine tune and troubleshoot some more.