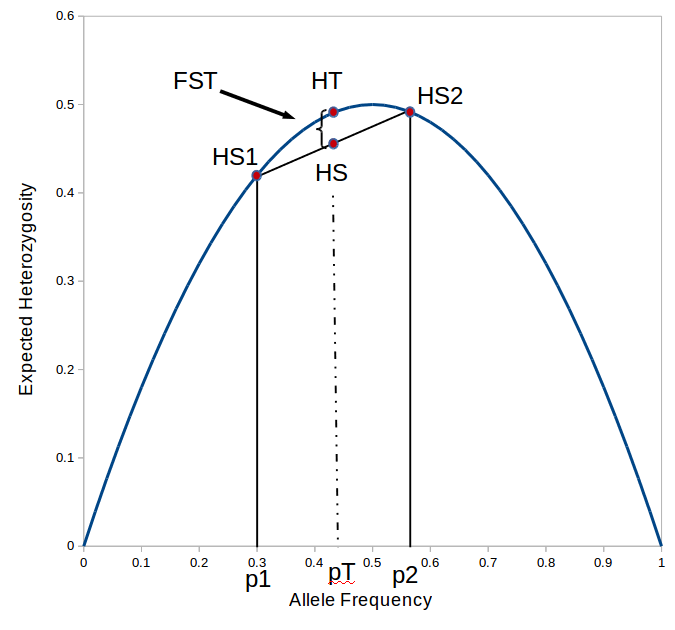
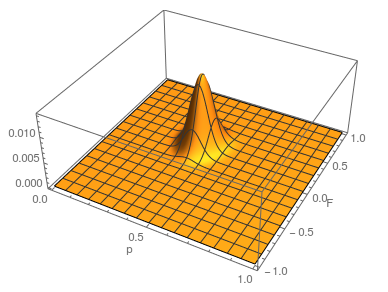
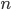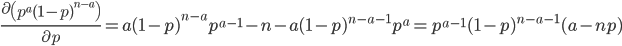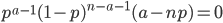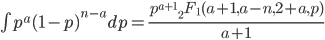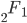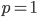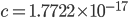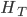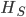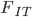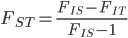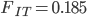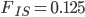Update: I converted the manuscript to TeX and submitted it to Arxiv:
http://arxiv.org/abs/1706.01710 (I also undid a couple of changes the reviewers wanted like changing Type 2 in the Appendix back to Type 0.)
Preface
The text in this post is from a "perspective" manuscript I was asked to write last year for a special issue of a journal focused on gene drive technologies. Long story short this manuscript was rejected and I see little recourse to submit it elsewhere for publication in a timely manner. However, I feel like many of the points in the manuscript should be made, at least in some form; so, I am posting it here.
Abstract
The Indo-Pacific region contains a unique mix of opportunities for the development and use of genetic-pest-management, gene-drive, and gene-drive-like technologies. Here I collectively refer to these technologies as Evolutionary Genetic Engineering (EGE). Indo-Pacific Islands have some of the world's highest rates of endemism and extinction—species and entire ecosystems are at risk. This threat to the natural world is coupled with the burden of human diseases, many of which are new and emerging or neglected tropical diseases. The same factors which have led to high rates of endemism also, in some ways, make this region an ideal testing ground for some types of EGE's. There is great potential for positive humanitarian, economic, and conservation applications of EGE's. However, these types of new technologies will be initially viewed from the perspective of the recent history of a loss of self determination, issues of social justice, and the testing of new technologies (e.g., biocontrol, agricultural, nuclear) in the Indo-Pacific—a region of the world that is still extensively colonized and controlled by Western Nations. Experience with successes and failures in related technologies suggests a path to move forward—a set of eight recommendations—to maximize the potential payoffs and minimize unintended negative effects of EGE's.
Introduction
The island Indo-Pacific is a large, important, unique, and unfortunately often overlooked region of the world. There is tremendous potential for the positive use of Evolutionary Genetic Engineering (EGE) in the region in both humanitarian and conservation applications. This potential stems from the regions geographic isolation, collection of infectious diseases, and species conservation urgencies. However, it would be a mistake to neglect the context of recent and ongoing political and social challenges in the region. Doing so is likely to generate a negative reaction that could inhibit the applications of promising emerging technologies. This context includes issues of colonialism, self determination, biocontrol, the testing of new technologies, and early experiences with genetically modified agricultural crops in the region. In this article I am focused on the island Indo-Pacific tropics, but also use examples from the broader region including India and Australia. I am also focusing on terrestrial applications of EGE’s. There are potential freshwater and marine applications, but this is less developed and goes beyond the scope of the current article.
In order to move forward in a way that does not sacrifice long term progress for short term convenience, we must accept that everyone has a role to play in shaping our technological future; this is not always easy to do when faced with confrontations and fundamental disagreements. To do this we must
- enhance communications and avoid a reluctance to provide more detailed information about new technologies or to be dismissive of inquiries.
- EGE applications should only be pursued if there is a genuine benefit to the local population (and if the people potentially affected generally agree that this is desirable rather than the decision being made externally), not in order to test new technologies in a “safe” manner or to avoid jurisdictional regulations.
- The potential benefits and risks of EGE's, along with the degree of uncertainty surrounding both, need to be unambiguously communicated.
- There needs to be a frank discussion of unintended side effects and the potential for misuse of the technology.
- Humanitarian goals need to be administered and controlled by humanitarian organizations while conservation goals need to be administered and under the control of conservation organizations (applicable to both governmental and non-governmental organizations); this is especially true in an international setting.
- Proactive research needs to be conducted and the data available to address common concerns about the possible ecological and health effects of EGE’s.
- It takes broad perspectives, beyond what any single person is capable of, to identify potential promises and pitfalls of the development and implementations of EGE's.
- Finally, a broad-based community discussion of, and direct involvement in, EGE development and applications should occur as early as possible. This will positively shape both the development and applications of the technology and help build a solid social foundation for future developments.
Potential Evolutionary Genetic Engineering Applications in the Island Indo-Pacific.
The Indo-Pacific spans half of the Earth’s circumference yet receives relatively less international focus. A revealing example is that this is the non-polar region that is most often divided on map projections of the world. This inattention is not simply due to a smaller population; the four most populous countries in the world (China, India, the US, and Indonesia) have territory and active interests in the region.
The island Indo-Pacific has some of the world’s highest rates of both species endemism (unique genetic diversity) and extinction (Vitousek 1988; Fleischer et al. 1998; Myers et al. 2000; Kier et al. 2009). Adaptive radiations of species here have served as prime examples of evolutionary biology (p. 380 Darwin 1845; Dobzhansky 1973). Extinctions in these species-rich regions are proceeding at an alarming rate (Pimm et al. 1995; Ganzhorn et al. 2001; Fonseca 2009; Loehle & Eschenbach 2012; Régnier et al. 2015) and this is predicted to be exacerbated by climate change (Benning et al. 2002; Mora et al. 2013). The region is in dire need of effective conservation strategies and potential Evolutionary Genetic Engineering (EGE) applications targeting introduced species and diseases have been proposed to establish effector genes refractory to introduced vectored disease (genetic modifications to block transmission of the disease) and genetic sterile insect techniques to suppress populations of invasive species (Clarke 2002; Wimmer 2005; Altrock et al. 2010; Esvelt et al. 2014; Reeves et al. 2014; Webber et al. 2015; see Sinkins and Gould 2006 and Gould et al. 2006 for review).
The island Indo-Pacific is also home to newly emerging and/or neglected tropical diseases that affect human health as well as economically important species. Vector borne human diseases in the region include chikungunya, dengue fever, Japanese encephalitis, lymphatic filariasis, malaria, plague and Rift Valley fever (Madagascar), schistosomiasis, scrub typhus, West Nile fever, and zika. Additionally there are diverse agricultural crop pests and diseases that impact food production across the region. A major goal of EGE development is to address human disease, and there is also potential for agricultural applications (Alphey 2002; Sinkins and Gould 2006; Gould 2008; Wimmer 2013; Esvelt et al. 2014; Champer et al. 2016).
When countries are listed by gross domestic product per capita it becomes apparent that the Indo-Pacific contains, in terms of national economic wealth, many of the poorest countries in the world. For example, the Comoros, Kiribati, Madagascar, Marshall Islands, Micronesia, Papua New Guinea, Solomon Islands, Tuvalu, and Vanuatu have an average per capita GDP of Intl.$2,387, approximately 1/8th of the world average, Intl.$18,872 (IMF 2016). This limits the resources available that these governments can apply to humanitarian and conservation interventions and suggests an enhanced value of international collaboration.
In many ways the terrestrial isolation that has led to the Indo-Pacific's tremendous biological diversity also makes the region ideal for some EGE applications. Suppressing or modifying non-native invasive species is an obvious place to start. However, what may be a pest in one location may be a highly valued or important ecological species in another (e.g., nopal Opuntia cacti are a highly valued component of Mexican cuisine and source of animal fodder while considered an invasive species pest in Australia—Cactoblastis cactorum has been used successfully as bio-control in Australia but is now threatening native Opuntia in the Americas, Zimmerman et al. 2004). Proper application of Type 1 and 3a EGE’s (Appendix A: Types of Evolutionary Genetic Engineering) can leave a species genetically unmodified within its native range, even with low levels of migration between islands or islands and continents (Altrock et al. 2010, 2011; Láruson & Reed 2016). The limited and discrete partitioning of land area of islands make 100% local genetic transformation or eradication of a species possible without resorting to type 3b or 4 EGE’s (Appendix A) and allows the application to proceed in a stepwise fashion across multiple islands using limited resources.
Colonialism, self determination, and the testing of new technologies
We have different perspectives depending on our experiences and social identities, and we are all-too-often not aware of how our individual perspective differs from others. In the middle of the abstract I used the following sentence, “The same factors which have led to high rates of endemism also, in some ways, make this region an ideal testing ground for some types of EGE's.” I chose the wording of this sentence carefully. What was your reaction? For many of the people reading this article the sentence seemed perfectly natural and flows into the ideas of the preceding and following sentences. However, for some readers the phrase “testing ground” is likely to stand out. Our reaction to this sentence is related to our perspective. For many who do not live in the island Indo-Pacific it is easy to see the region as something external to our daily lives and more disposable for testing and experimenting. In contrast, for some the Indo-Pacific represents home, family, work, and is also fundamentally connected to a cultural identity. I ask readers to construct your own sentence connecting a place that is highly valued to you personally (your hometown, where you live now, or a place of historical, religious, or cultural importance) with a “testing ground” of a new potentially powerful technology with its own set of concerns and unknowns.
The Indo-Pacific has a long and continuing history of a loss of self determination and sovereignty. The UN Special Committee on Decolonization lists American Samoa, French Polynesia, Guam, New Caledonia, Pitcairn, and Tokelau as Non-Self-Governing-Territories. The total number of ongoing sovereignty disputes encompasses many more islands and regions too extensive to list here. Colonization includes the establishment of extensive military bases and use of the islands for tests of nuclear, biological, and chemical warfare technologies—and these were not limited to a few isolated incidents, for example hundreds of nuclear weapons tests were conducted by France in Mururoa and Fangataufa Atolls, by the United Kingdom in South Australia, Montebello, and Kiritimati Islands, by the United States in Pikinni (Bikini), Ānewetak (Enewetak), Johnston (Kalama) Atolls, and Kiritimati. This history of military testing, non-military testing of new technologies (e.g., disastrous attempts at classical biological control by introducing new species, e.g., Howarth 1983; Clarke et al. 1984; Henneman & Memmott 2001; Messing & Wright 2006; Hays and Conant 2007; Parry 2009), and colonization in the region can severely inhibit international biological research and potential applications including EGE’s.
There is a case study that deserves special mention within the context of EGE’s in the Indo-Pacific, especially in the context of international programs and applications of mosquito genetic engineering. From 1969–1975 the World Health Organization (WHO) collaborated with the US Public Health Service (PHS) and the Indian Council of Medical Research (ICMR) to establish a Genetic Control of Mosquitoes Research Unit (GCMRU) in India; this was financially supported by the Government of India, US PL-480 funds, and the CDC (AEND 1975a). The GCMRU was studying and implementing mosquito control technologies including the release of sterilized individuals. What appears to have started with concerns about a carcinogen (thiotepa) being added to well water in the village of Pochanpur without public or government consultation got caught up in politics (AEND 1975b; Hanlon 1975), with widespread accusations in the media and later by the Government of India, and grew into a political disaster with suspicions that the US military was using India to test methods of biological warfare using mosquitoes (Sehgal 1974; Anonymous 1975; AEND 1974; AEND 1975b; Hanlon 1975; Anonymous 1976; Powell and Jayaremen 2002). The addition of thiotepa to village water has been denied by WHO (Tomiche 1975), but publications preceding the accusations suggest this may have happened (pp. 85, 87, Pal 1974)—and therein lies one problem. There was a lack of clear unambiguous communication from the beginning. Furthermore, PHS did have military connections and shared materials and information with the US military (Langer 1967; Treaster 1975). The US military did conduct chemical and biological tests in the Indo-Pacific; this included the release of mosquitoes off the coast of Baker Island (“Magic Sword” 1965), the release of Bacillus globigii in Oʻahu (“Big Tom” 1965), shelling sarin nerve agent in Waiākea Forest Reserve, Hawaiʻi (“Red Oak” 1967), and the dispersal of Staphylococcus aureus enterotoxin type B over Ānewetak (Enewetak) Atoll (“DTS Test 68-50” 1968). However, in all likelihood there was no military or biological warfare connections with the GCMRU (WHO 1976; Powell and Jayaremen 2002). Covert transfer of US funds to keep GCMRU going was briefly discussed with WHO (AEND 1975c; SSWDC 1975) but this was considered too risky and the US suspended funding the project. Despite denials by WHO (Tomiche 1975), the GCMRU, which was planned to extend at least until 1978, was forced to shut down prematurely in 1975 (AEND 1975d) and the project was deemed a failure (Curtis 2007).
What can be learned from this?
- There was a clear lack of communication resulting from a reluctance of either the WHO or the US to engage the media and comment on the allegations (AEND 1974; AEND 1975e; Anonymous 1975; Tomiche 1975). This was unfortunate as it, perhaps rationally, fueled suspicions. The public perception of public perception may differ from public perception—the individual perception of public opinion is influenced by a range of factors and may not be an accurate reflection of commonly held attitudes (e.g., Mutz 1989). The idea that providing more information would undermine support conflicts with recent results that show the more informed people are of the release of genetically modified mosquitoes the more supportive they become; however, a great deal of public engagement has to be accomplished, especially for women, minorities, and people with lower education levels and lower household incomes (Ernst et al. 2015; Kolopack et al. 2015).
- There was a perception that these experiments would not have been permitted in Western countries and that India was being used as a testing ground (Anonymous 1975; Raghavan and Jayaraman 1975). Knowledge that a technology has been effective in other countries is one factor associated with strong public support (Ernst et al. 2015). It is unfortunate that prior programs in the US, Myanmar/Burma, Tanzania, Western Africa, and France were not communicated to the Indian press (Laven 1972; WHO 1976; Curtis 2007). The perception that an international project is being conducted to avoid home country regulation should certainly be (truthfully) avoided.
- The potential benefits of the project to the people of India was unclear (AEND 1974; Anonymous 1975). This is perhaps most tragic of all. India suffers from mosquito vectored dengue, malaria, Japanese encephalitis, Chikungunya, and lymphatic filariasis (Sharma 2015). While a balance must be struck to not over-promise results that may not be realized, the goals and potential benefits of EGE applications must also be clearly advertised.
- There was a lack of an a priori open and frank discussion about possible misuse of the technology (Hanlon 1975). While any technology can be potentially misused by individuals or organizations, a nation's government, and especially its military, has non-humanitarian and non-conservation priorities that can potentially conflict with the goals of humanitarian and conservation projects. Regardless of the existence of an actual conflict, the perception of possible conflict does exist, which can undermine credibility (Serafino et al. 2008; Charny 2013). Fortunately today this is widely recognized and the 1978 UN ENMOD treaty (http://www.un-documents.net/enmod.htm) may prevent, depending on interpretation, military involvement in EGE technologies except perhaps for some limited applications of type 1 and 2 systems (Appendix A). The ENMOD treaty states that “Each State Party to this Convention undertakes not to engage in military … environmental modification techniques having widespread, long-lasting or severe effects … the term "environmental modification techniques" refers to any technique for changing—through the deliberate manipulation of natural processes—the dynamics, composition or structure of the Earth, including its biota …“ However, there is still a need for an open discussion about potential malicious uses, and military involvement with EGE projects should be avoided in order to encourage international trust and cooperation.
In response, Dr. B. D. Nagchaudhuri’s, physicist and scientific adviser to the Indian Ministry of Defense, recommendations were “(A) that research proposals and projects are available to the public; and (B) that pertinent records contain clear statements as to why the objective is important, what is the [Government of India's] interest, and what is the [United States Government’s] interest” (AEND 1975f) and “ministry officials must be alerted to any sensitive problems by the technical experts involved”; also, that “each collaborative project should also be approved at the ministerial or secretary level of the ministry under which the project would fall i.e. health projects - Ministry of Health, Agricultural Projects - Ministry of Agriculture, … This should also hold true whether on the Indian side or the US side” (AEND 1975g). Dr. Hanlon recommends “At the very least, there should be an open discussion of the [biological warfare] potential of such projects before they begin, so that countries can make informed choices” (p. 103, Hanlon 1975).
There is a value to compartmentalizing different aspects of a government’s actions. It seems almost self evident that funds for research are best spent by research agencies, funds for health are best spent in agencies focused on health, funds for conservation are best spent by agencies trained in and focused on conversation. Even if there were sufficient funding and resources we would not want the EPA (Environmental Protection Agency) or DOH (Department of Health) carrying out military actions; the converse is also true. We don’t want to rely on our military to carry out conservation, human health, or humanitarian actions when there are other agencies, without conflicting priorities, that can and should be doing this (Serafino et al. 2008; Charny 2013). The author has discussed EGE's and the ENMOD treaty in person with current and former members of DARPA, a research branch of the military with an interest in EGE's, and has been told that the military has to carry out high risk (in the sense of new and experimental) research because NSF (National Science Foundation) and NIH (National Institutes of Health) cannot. I disagree. Research agencies can and should also be funding higher risk, higher payoff research instead of abdicating this role to the military—and to avoid the kinds of conflicts suggested in the WHO experience in India. This is not done in the US because of historical inertia and objectively unbalanced federal budget allocations (a Department of Defense, DOD, estimated research budget of $66 billion versus $29 billion for NIH and $6 billion for NSF in FY2015; Hourihan and Parkes 2016). Reallocating civilian research funds to civilian agencies would also free up the military to focus on military actions and capabilities.
Recent experiences with GMO’s in the Indo-Pacific
EGE’s are likely to be initially framed in terms of the GMO (Genetically Modified Organism) crop debate (Knols et al. 2007). Within Hawaiʻi, Rainbow Papaya and GMO Taro serve as contrasting examples of the interaction between social acceptance, development, and deployment of new technologies. Carica papaya was not grown in Hawaiʻi until after European contact in 1778. The papaya industry in Hawaiʻi was devastated in the 1990’s by the ringspot virus. A genetically engineered “rainbow” papaya resistant to ringspot infection was developed at Cornell University by Dr. D. Gonsalves (Ferreria et al. 2002) who was originally from Hawaiʻi. While GMO papaya is not without controversy (e.g., Harmon 2014; see Hofschneider 2016 for recent legal developments in Hawaiʻi) it is credited with rescuing the industry and is de facto widely adopted in Hawaiʻi today (e.g., Kallis 2013).
Colocasia esculenta (Taro or Kalo in Hawaiian) was brought to Hawaiʻi by the ancient Polynesians. A wide range of Kalo varieties have had a central role in traditional Hawaiian culture as a staple food crop and continues to be economically important (Whitney et al. 1939; Fleming 1994). Furthermore, Kalo is literally the brother of humans (Hāloa) in the Hawaiian creation tradition and words for family and relationships also refer to parts of the plant (Kahumoku 1980). Taro leaf blight (Phytophthora colocasiae) was introduced to Hawaiʻi in the 1900’s and has significantly impacted Kalo (Nelson et al. 2011). Work at the University of Hawaiʻi was begun to to breed resistant varieties which resulted in patents in 2002. Separately a Chinese variety of Taro was genetically modified from 2001 to 2006 with a gene from wheat to be resistant to leaf blight. This resulted in widespread public outrage and large protest rallies in 2006 that resulted in the university relinquishing its patents and issuing an indefinite moratorium on the genetic engineering of Hawaiian Kalo (Ritte and Freese 2006; CTAHR 2009).
With these cases in mind consider a potential EGE project. Culex mosquitoes were introduced to Hawaiʻi in the mid 1800’s. They vector Plasmodium relictum which is responsible for avian malaria. Many Hawaiian forest bird species, important in traditional Hawaiian culture (e.g., ʻahu ʻula, mahiole, and in Hawaiian religion), have no immunity or tolerance to P. relictum and have become extinct, with many currently threatened, as a result (Warner 1968). These two previous contrasting examples suggest that genetically modifying non-native mosquitoes to reduce the frequency of avian malaria is much more socially acceptable than the reverse: genetically modifying native Hawaiian birds to be resistant to infection by Plasmodium (although it would be worth conducting the relevant public surveys to determine this). Also, doing the research locally in Hawaiʻi is not necessarily an advantage in terms of securing broad local public support, buy-in, and acceptance (however, it is an advantage in terms of engaging the public). These are aspects that might not initially be appreciated by scientists designing EGE technologies.
On a broader scale across the Indo-Pacific, consider the cases of golden rice and Bt-cotton. Rice (Oryza sativa) is a staple crop for a large segment of the population across the Indo-Pacific. A major nutritional shortcoming of rice is the lack of beta-carotene that can be metabolized into vitamin A, which in many of these populations is de facto not simply rectified by supplementing with additional food sources. This unfortunate situation leads to blindness and the deaths of over half a million people a year. To address this, rice has been engineered since 2000 with DNA sequences from other plants to produce bio-available beta-carotene (Ye et al. 2000; Paine et al. 2005; Tang et al. 2009). This “golden rice” has also been the target of a great deal of controversy, protest, and misinformation (e.g., Dobson 2000; Potrykus 2001; Enserink 2008; Lynas 2013). Much of this protest originates in the Western world where ironically we have a wide range of nutritional supplements added to our food including vitamin D in milk, calcium in orange juice, niacin and folic acid in bread, iodine in salt, and fluoride in drinking water. One question to ask ourselves is, why is it so easy to add all of these supplements to our food supply, not to mention widespread adoption of genetically modified corn, soybeans, cotton, potatoes, sugar beets, etc., in parts of the West, when providing vitamin A in the form of Golden Rice for much of the world’s population is still not approved and remains in a testing phase well over a decade later?
Bt-cotton, which has received less attention in the media, provides a contrasting case to golden rice where a GM crop has been embraced in the Indo-Pacific and this has been in large part driven by local buy-in. Bt-cotton is engineered to produce a naturally occurring insecticide from a bacteria (Bacillus thuringiensis). The intention is to kill larvae of the cotton bollworm (Helicoverpa armigera). A seed company in India led by Dr. D. B. Desai began selling “Navbharat 151” seed in 1998 with the claim that the plants did not have to be sprayed with pesticides for bollworm. This proved to be the case during a large bollworm outbreak in Gujarat in 2001, which raised questions. It was found that Navbharat 151 plants had a genetic modification created by Monsanto. The Indian government filed criminal charges against Dr. Desai, ordered the seed destroyed, and 4,000 hectares of planted fields burned. Thousands of farmers rallied to support Dr. Desai and block burning the fields; the Gujarat government refused to carry out the order; the recall was canceled, and some farmers saved their own seed for replanting. The opposite of concerns about using India as a testing ground as discussed in the WHO mosquito project of the 1970's (point 2 above) were expressed: 'How can something made in the United States, many of them wonder aloud, be unsafe in India? “I think they grow it in China and other countries,” says Kalidas Patel, who grew Navbharat cotton in Gujarat' (McGray 2002). Later Monsanto was granted a license to market Bt-cotton in India and in all likelihood the prior experience with Navbharat 151 promoted public buy-in (Menon 2001; McGray 2002). In recent years Bt-cotton is widely adopted, approximately 90% of the cotton grown in India, and a black market for Bt-cotton seeds also appears to be thriving (e.g., Kathage and Qaim 2012; Nemana 2012). However, this is in no way a simple matter and debates regarding Bt-cotton, Monsanto, and regulation continue (e.g., Anonymous 2016; Basheer 2016). Regardless, the support among Indian farmers for Bt-cotton stands in stark contrast to the protests over golden rice being planted in test beds in the Philippines (Lynas 2013). The cause of the difference between these experiences is hard to isolate and a large number of idiosyncratic effects likely contribute including the pivotal actions of a few or a single individual. However, the effects of local buy-in, combined with local access to technologies, and first hand experience with these technologies, should not be ignored.
Finally, concerns about ecological effects of EGE’s are associated with strong opposition to the technology (Ernst et al. 2015). There are also questions of possible, but unlikely, bioaccumulation of toxic proteins and allergenicity (Curtis 2007; Reeves et al. 2012). In addition to the four guidelines in the previous section, despite limited time and funding, we should conduct the work to have the data on hand to address these questions to the public (Curtis 2007).
Everyone has a role to play
We live in a world that is often overly self-polarizing. I am a geneticist; I entered this field because of personal interest, excitement, and challenges of the promise and potential of genetics. Unintentionally, this has become a part of my identity. When I was first exposed to protests over genetic technology it was all too easy to feel that it was also a personal attack. This is nested within the context of broader anti-scientific popular views related to climate change, evolution, renewable energy, vaccinations, etc. The natural reaction is to reflexively move in the opposite direction and argue that genetic technologies are safe, protesters don't understand the issues, etc. and be overly dismissive; a position that I may not have had initially. The difficult but essential step for growth is to try to find a middle ground and synthesize a path forward (see also NPR 2013). Right or wrong, no single perspective can do this on its own and, because of our perspectives, we are often blind to potential issues apparent to other people. It is easier to see a potential risk if you are looking for a risk instead of working toward developing a desired application of a new technology. For example, the potential of allergic reactions to genetic modifications are real and not to be dismissed (e.g., Nordlee et al. 1996), and many crops have a strong cultural significance that many people may not be aware of such as Kalo in Hawaiʻi, discussed above, or maize in Chiapas (Bellon and Brush 1994; Peralez et al. 2005; Brush and Peralez 2007). As geneticists we are in a unique position to be able to critically assess potential benefits and risks, once we perceive them, of genetic technology from a scientific perspective. It is our responsibility to embrace and communicate this rather than contributing to destructive polarization. However, it is not our job to be overly encompassing and give equal weight to all objections; we also must be willing to rationally disagree when we reason this to be the case. For example, despite claims to the contrary (Séralini et al. 2012), there is no scientific evidence that herbicide resistant maize is carcinogenic. There is a great deal of misinformation and misconceptions surrounding who would or would not benefit, and to what degree, from golden rice (Harmon 2013). Attitudes regarding GMO's are divisive, some are not based on factual evidence and can be labeled as irrational although this quickly gets complex (Stone 2010; Lynas 2013; Blancke et al. 2015; Hicks 2015); regardless, the GMO debate will continue to prove a rich subject for the analysis of the dynamics of politics, the media, framing effects, confirmation bias, social identity, information cascades, etc., for many years to come.
An area that can benefit from improvement is to incorporate this synthesis earlier into the research and development process. If individuals with different perspectives were able to directly participate in the design of a new technology, they could shape the direction in which it develops towards an outcome that might be more desirable and socially acceptable. (Recall the effect of personal experience with Bt-cotton and its adoption in India.) Often the way development of a new technology works is in incremental steps of design, troubleshooting, and research funding, to consultation and approval from regulatory agencies, to building the logistics of application and deployment. Public consultation and asking for acceptance occurs only at the end of the day, when many steps have been cast and it is more difficult and time consuming to make fundamental revisions. One possibility is to include grant support for individuals from the social sciences to be “embedded” in a biological laboratory in order to fully participate in a laboratory’s research and conduct their own research about social attitudes, context, communication, perceptions, etc., both in its own right and as a bidirectional conduit to facilitate communication, public guidance, and knowledge transfer in the development of EGE technologies (see Kolopack et al. 2015 for a highly effective example of community engagement albeit not exactly in the same form that I am proposing here). The local community can directly participate in the development of a new technology, possibly facilitating progress in a direction that is unanticipated by the researchers, funding, and regulatory agencies, but one that results in a greater positive potential being realized at the end of the day.
Conclusion
This Indo-Pacific is geographically isolated, under a burden of infectious diseases, and is in dire need of protection of its natural heritage. This creates an opportunity for positive, highly valued, effective applications of EGE's. However, it would be a mistake to ignore the history and social realities thought the region. To reiterate the eight points from the introduction that have been expanded upon through this article:
- There is a need to enhance and engage communications in all directions.
- EGE applications should only be pursued if there is a genuine benefit to, and buy-in from, the local population.
- The potential benefits and risks of EGE's need to be unambiguously communicated.
- There needs to be a clear unambiguous discussion of unintended side effects and potential misuses of the technology.
- Humanitarian goals need to be administered and controlled by humanitarian organizations and conservation goals need to be administered and under the control of conservation organizations.
- Proactive research needs to be conducted and the data available to address common concerns.
- It takes broad perspectives to broadly identify potential promises and pitfalls of EGE's.
- An early broad-based community discussion of, and involvement in, EGE development and applications should occur.
Finally, no matter how “new” a technology or situation seems, there is still much to be learned from history.
Appendix A: Types of Evolutionary Genetic Engineering
An important concept that cannot be over-emphasized is the diverse types of EGE's and their predicted effects. At the risk of oversimplification, here are four main types with an important boundary between them.
- Type 1
- Deleterious EGE's that are designed to be transient and removed from the population. Examples of type 1 include the “killer-rescue” system (Gould et al. 2008), genetic sterile insect technique (Horn and Wimmer 2003), and Wolbachia in cytoplasmic incompatibility population suppression applications (Laven 1967; Knipling et al. 1968). These may persist in the wild for a shorter period of time than type 2 EGE’s.
- Type 2
- Generic genetic modifications not designed to change in frequency over time using evolutionary principles. In general these are expected to either drift neutrally (if there is little to no effect) or be removed by natural selection. For example fluorescent proteins are often used to mark and keep track of genetic inserts; however, these proteins can have toxic effects (e.g., Liu et al. 1999; Devgan et al. 2004; Shaner et al. 2004, 2005). This tends to reduce an organism’s fitness and these modifications are not expected to persist in the wild over many generations.
- Type 3
- Threshold EGE’s that cannot increase in frequency when very rare but can increase in frequency and persist indefinitely once a critical frequency point is passed.
- Type 3a
- Thresholds that are above a frequency of one half. These include chromosomal rearrangements (Foster et al. 1972), haploinsufficient induced underdominance (Reeves et al. 2014) and possibly some forms of maternal-effect underdominance (Akbari et al. 2013).
- Type 3b
- Thresholds that are below a frequency of one half. This includes Wolbachia (Hoffman et al. 2011), some forms of maternal-effect underdominance (Akbari et al. 2013), and some theoretical systems (Davis et al. 2001).
- Type 4
- Unconditionally driving EGE’s that can invade a population from arbitrarily low frequencies. These include Medea (Chen et al. 2007), homing endonucleases (Windbichler et al. 2011), transposable elements (Caraeto et al. 1997), meiotic drive (Cha et al. 2006) and some types of CRISPR systems (Gantz and Bier 2015; DiCarlo et al. 2015; Hammond et al. 2016).
In one perspective, the most important distinction is the boundary between 3a and 3b. This predicts what will happen without human intervention (releases of modified or unmodified individuals) among multiple populations within a species due to the forces of migration and selection (Barton and Turelli 2011). Type 1—3a will tend to reduce in range and disappear (although this may take many generations for 2 and 3a) while type 3b and 4 will tend to spread and become more established (and this may occur in a small number of generations for type 4) with the concern that once widespread enough this may be irreversible. While type 3a systems might be considered "gene drive" in a broad sense the term is probably more accurate to describe type 3b and especially type 4 systems (gene drive in the strong sense). The boundary between 3a and 3b represents a balance between ease of transformation of a population and reversibility back to a transformation free state—a balance between safety and efficiency.
Some natural EGE systems in the type 4 category have been shown to be capable of moving across subspecies and species boundaries, rapidly spreading worldwide, and lowering the average fitness of a species (e.g., Eanes et al. 1988; Morita et al. 1982; Hill et al. 2016). The concern of this possibly happening due to artificial genetic engineering is not a new one (Gould et al. 2006). For example, fully functioning transposable elements have been introduced into various new species in the lab (e.g., Brennan et al. 1984; Daniels et al. 1989), and this is often done with little to no discussion of possible escape. Fortunately there are methods of building in safeguards to minimize the chance of unintended spread in the wild (Dafa’alla et al. 2006; Gokhale et al. 2014).
Additionally, it is important to keep in mind the (sometimes unexpected) effects of mutations and selection that can change the dynamics of EGE’s. For example, Y chromosome meiotic drive can be quickly suppressed by sex chromosome aneuploidy (Lyttle 1981). Arthropod species have been observed to rapidly evolve to suppress some effects of Wolbachia (Charlat et al. 2007). Type 2 EGE’s may drift at some frequency in a population by unintended contamination (e.g., Gonzales et al. 2012; Xiao et al. 2016); one concern that goes beyond this is that genetically engineered disease resistance may be adaptive, if infection by the disease has a large enough fitness cost, and the type 2 EGE may deterministically increase in frequency in the wild, essentially becoming a type 4 EGE (although to date there are not clear examples of this, e.g., Fuchs et al. 2004). Some of these unexpected effects can be detected in laboratory experiments and incorporated into the design and predictions of the EGE.
It is already a challenge to filter out misinformation and misconceptions regarding genetic modifications. The author realizes that this adds another challenge; however, the fact is there are various types of EGE's with a range of predicted effects regarding how well they can be established in the wild and how reversible they are. It is appropriate, if possible, for these dynamics to be considered and to interact with regulatory approval and public acceptance (Harmon 2014).
References
Alphey, L. (2002). Re-engineering the sterile insect technique. Insect Biochemistry and Molecular Biology, 32(10), 1243-1247.
Altrock, P. M., Traulsen, A., Reeves, R. G., & Reed, F. A. (2010). Using underdominance to bi-stably transform local populations. Journal of Theoretical Biology, 267(1), 62–75.
Altrock, P. M., Traulsen, A., & Reed, F. A. (2011). Stability properties of underdominance in finite subdivided populations. PLoS Computational Biology, 7(11).
Akbari, O. S., Matzen, K. D., Marshall, J. M., Huang, H., Ward, C. M., & Hay, B. A. (2013). A synthetic gene drive system for local, reversible modification and suppression of insect populations. Current Biology, 23(8), 671-677.
AEND: American Embassy New Delhi. (1974). NEW DE 10039: Indian press articles on WHO biogenetic mosquito control project and migratory bird project. Declassified/Released US Department of State EO Systematic Review, (July). Retrieved from https://wikileaks.org/plusd/cables/1974NEWDE10039_b.html
AEND: American Embassy New Delhi. (1975a). NEW DE 08068: WHO mosquito genetic project. Declassified/Released US Department of State EO Systematic Review, (June). Retrieved from https://wikileaks.org/plusd/cables/1975NEWDE08068_b.html
AEND: American Embassy New Delhi. (1975b). New DE 06070: US and WHO again linked with germ warfare activities in India. Declassified/Released US Department of State EO Systematic Review. Retrieved from https://wikileaks.org/plusd/cables/1975NEWDE06070_b.html
AEND: American Embassy New Delhi. (1975c). NEW DE 14272: WHO mosquito genetic control project. Declassified/Released US Department of State EO Systematic Review. Retrieved from https://wikileaks.org/plusd/cables/1975NEWDE14272_b.html
AEND: American Embassy New Delhi. (1975d). NEW DE 09077: Genetic control of mosquitoes. Declassified/Released US Department of State EO Systematic Review. Retrieved from https://wikileaks.org/plusd/cables/1975NEWDE12250_b.html
AEND: American Embassy New Delhi. (1975e). NEW DE 12250: Refutation of germ warfare charges. Declassified/Released US Department of State EO Systematic Review. Retrieved from https://wikileaks.org/plusd/cables/1975NEWDE12250_b.html
AEND: American Embassy New Delhi. (1975f). NEW DE 09023: US-GOI Research Projects. Declassified/Released US Department of State EO Systematic Review, (July). Retrieved from https://wikileaks.org/plusd/cables/1975NEWDE09023_b.html
AEND: American Embassy New Delhi. (1975g). NEW DE 12522: Indo-US joint commission meeting in October -- working draft of minutes. Declassified/Released US Department of State EO Systematic Review. Retrieved from https://wikileaks.org/plusd/cables/1975NEWDE12522_b.html
Anonymous. (1975). Oh, New Delhi; oh, Geneva. Nature, 256, 355–357.
Anonymous (1976) Indian attack on foreign research funding. New Scientist, 20 May, 396.
Anonymous (2016) Monsanto kicked out for giving farmers raw deal. The Times of India. Published online http://timesofindia.indiatimes.com/city/mumbai/monsanto-kicked-out-for-giving-farmers-raw-deal/articleshow/55991161.cms
Barton, N. H., & Turelli, M. (2011). Spatial waves of advance with bistable dynamics: cytoplasmic and genetic analogues of Allee effects. The American Naturalist, 178(3), E48-E75.
Basheer, S. (2016). The battle over Bt cotton. The Hindu. Published online http://www.thehindu.com/opinion/op-ed/The-battle-over-Bt-cotton/article15424211.ece
Bellon, M. R., & Brush, S. B. (1994). Keepers of maize in Chiapas, Mexico. Economic Botany, 48(2), 196-209.
Benning, T. L., LaPointe, D., Atkinson, C. T., & Vitousek, P. M. (2002). Interactions of climate change with biological invasions and land use in the Hawaiian Islands: modeling the fate of endemic birds using a geographic information system. Proceedings of the National Academy of Sciences, 99(22), 14246-14249.
Blancke, S., Van Breusegem, F., De Jaeger, G., Braeckman, J., & Van Montagu, M. (2015). Fatal attraction: the intuitive appeal of GMO opposition. Trends in Plant Science, 20(7), 414-418.
Brennan, M. D., Rowan, R. G., & Dickinson, W. J. (1984). Introduction of a functional P element into the germ-line of Drosophila hawaiiensis. Cell, 38(1), 147-151.
Brush, S. B., & Perales, H. R. (2007). A maize landscape: Ethnicity and agro-biodiversity in Chiapas Mexico. Agriculture, Ecosystems & Environment, 121(3), 211-221.
Carareto, C. M., Kim, W., Wojciechowski, M. F., O'grady, P., Prokchorova, A. V., Silva, J. C., & Kidwell, M. G. (1997). Testing transposable elements as genetic drive mechanisms using Drosophila P element constructs as a model system. Genetica, 101(1), 13-33.
Cha, S. J., Mori, A., Chadee, D. D., & Severson, D. W. (2006). Cage trials using an endogenous meiotic drive gene in the mosquito Aedes aegypti to promote population replacement. The American Journal of Tropical Medicine and Hygiene, 74(1), 62-68.
Champer, J., Buchman, A., & Akbari, O. S. (2016). Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nature Reviews Genetics, 17(3), 146-159.
Charlat, S., Hornett, E. A., Fullard, J. H., Davies, N., Roderick, G. K., Wedell, N., & Hurst, G. D. (2007). Extraordinary flux in sex ratio. Science, 317(5835), 214-214.
Charny, J. R. (2013). The U.S. Military's Expanding Role in Foreign Assistance. InterAction Policy Brief. Published Online https://www.interaction.org/files/FABB%202013_Sec16_NGOAndMilitaryRelations.pdf
Chen, C. H., Huang, H., Ward, C. M., Su, J. T., Schaeffer, L. V., Guo, M., & Hay, B. A. (2007). A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science, 316(5824), 597-600.
Clarke, B., Murray, J., & Johnson, M. S. (1984). The extinction of endemic species by a program of biological control. Pacific Science, 38(2), 97-104.
Clarke, T. (2002). Mosquitoes minus malaria. Nature, 419(6906), 429-430.
CTAHR: College of Tropical Agriculture and Human Resources (2009). CTAHR and Taro. Published online http://www.ctahr.hawaii.edu/oc/freepubs/pdf/CTAHR_and_taro.pdf
Curtis, C. F., 2007 Destruction in the 1970s of a research unit in India on mosquito control by sterile male release and a warning for the future. Antenna 31(4): 214-216.
Dafa'alla, T. H., Condon, G. C., Condon, K. C., Phillips, C. E., Morrison, N. I., Jin, L., ... & Alphey, L. (2006). Transposon-free insertions for insect genetic engineering. Nature Biotechnology, 24(7), 820-821.
Daniels, S. B., Chovnick, A., & Kidwell, M. G. (1989). Hybrid dysgenesis in Drosophila simulans lines transformed with autonomous P elements. Genetics, 121(2), 281-291.
Darwin, C. R. 1845. Journal of researches into the natural history and geology of the countries visited during the voyage of H.M.S. Beagle round the world, under the Command of Capt. Fitz Roy, R.N. 2d edition. London: John Murray.
Davis, S., Bax, N., & Grewe, P. (2001). Engineered underdominance allows efficient and economical introgression of traits into pest populations. Journal of Theoretical Biology, 212(1), 83-98.
Devgan, V., Rao, M. R., & Seshagiri, P. B. (2004). Impact of embryonic expression of enhanced green fluorescent protein on early mouse development. Biochemical and Biophysical Research Communications, 313(4), 1030-1036.
DiCarlo, J. E., Chavez, A., Dietz, S. L., Esvelt, K. M., & Church, G. M. (2015). Safeguarding CRISPR-Cas9 gene drives in yeast. Nature Biotechnology 33: 1250–1255
Dobson, R. (2000), Royalty-free licenses for genetically modified rice made available to developing countries, Bulletin of the World Health Organisation, 78 (10): 1281
Dobzhansky, T. (1973). Nothing in Biology Makes Sense Except in the Light of Evolution. The American Biology Teacher, 35(3), 125–129.
Eanes, W. F., Wesley, C., Hey, J., Houle, D., & Ajioka, J. W. (1988). The fitness consequences of P element insertion in Drosophila melanogaster. Genetical Research, 52(01), 17-26.
Enserink, M. (2008). Tough Lessons From Golden Rice. Science 320 (5875): 468–471
Ernst, K. C., S. Haenchen, K. Dickenson, M. S. Doyle, K. Walker, A. J. Monagen, and M. H. Hayden. 2015. Awareness and support of release of genetically modified “sterile” mosquitoes, Key West, Florida, USA. Emerging Infectious Diseases 21(2): 320-324.
Esvelt, K. M., Smidler, A. L., Catteruccia, F., & Church, G. M. (2014). Concerning RNA-guided gene drives for the alteration of wild populations. Elife, 3, e03401.
Ferreira, S. A., Pitz, K. Y., Manshardt, R., Zee, F., Fitch, M., & Gonsalves, D. (2002). Virus Coat Protein Transgenic Papaya Provides Practical Control of Papaya ringspot virus in Hawaii. Plant Disease, (February), 101–105.
Fleischer, R. C., McIntosh, C. E., & Tarr, C. L. (1998). Evolution on a volcanic conveyor belt: using phylogeographic reconstructions and K–Ar‐based ages of the Hawaiian Islands to estimate molecular evolutionary rates. Molecular Ecology, 7(4), 533-545.
Fleming, K. (1994). The Economics of Commercial Wetland Taro Production in Hawaii. Agribusiness, 7, 1–6.
Fonseca, C. R. (2009). The silent mass extinction of insect herbivores in biodiversity hotspots. Conservation Biology, 23(6), 1507-1515.
Foster, G. G., Whitten, M. J., Prout, T., & Gill, R. (1972). Chromosome rearrangements for the control of insect pests. Science, 176(4037), 875–880.
Fuchs, M., Chirco, E. M., & Gonsalves, D. (2004). Movement of coat protein genes from a commercial virus-resistant transgenic squash into a wild relative. Environmental Biosafety Research, 3(1), 5-16.
Gantz, V. M., & Bier, E. (2015). The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science, 348(6233), 442-444.
Ganzhorn, J. U., Lowry, P. P., Schatz, G. E., & Sommer, S. (2001). The biodiversity of Madagascar: one of the world’s hottest hotspots on its way out. Oryx, 35(4), 346-348.
Gokhale, C. S., Reeves, R. G., & Reed, F. A. (2014). Dynamics of a combined medea-underdominant population transformation system. BMC Evolutionary Biology, 14(1), 1.
Gonsalves, D., Gonsalves, C., Carr, J., Tripathi, S., Matsumoto, T., Suzuki, J., ... & Pitz, K. (2012). Assaying for pollen drift from transgenic ‘Rainbow’ to nontransgenic ‘Kapoho’ papaya under commercial and experimental field conditions in Hawaii. Tropical Plant Biology, 5(2), 153-160.
Gould, F. (2008). Broadening the application of evolutionarily based genetic pest management. Evolution, 62(2), 500-510.
Gould, F., Magori, K., & Huang, Y. (2006). Genetic Strategies for Controlling Mosquito-Borne Diseases. American Scientist, 94, 238–246.
Gould, F., Huang, Y., Legros, M., & Lloyd, A. L. (2008). A Killer–Rescue system for self-limiting gene drive of anti-pathogen constructs. Proceedings of the Royal Society of London B: Biological Sciences, 275(1653), 2823-2829.
Hammond, A., Galizi, R., Kyrou, K., Simoni, A., Siniscalchi, C., Katsanos, D., ... & Burt, A. (2016). A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nature Biotechnology, 34(1), 78-83.
Hanlon, J. 1975. Germ-war allegations force WHO out of Indian mosquito project. New Scientist 68(970, 9 Oct) 102-103
Harmon, A. 2013. Golden Rice: Lifesaver? The New York Times, August 25, 2013, New York Edition, page SR1. Available online http://www.nytimes.com/2013/08/25/sunday-review/golden-rice-lifesaver.html
Harmon, A. 2014. On Hawaii, a Lonely Quest for Fact. The New York Times, January 5, 2014, New York Edition, page A1. Available online https://www.nytimes.com/2014/01/05/us/on-hawaii-a-lonely-quest-for-facts-about-gmos.html
Hays, W. S., & Conant, S. (2007). Biology and impacts of Pacific Island invasive species. 1. A worldwide review of effects of the small Indian mongoose, Herpestes javanicus (Carnivora: Herpestidae). Pacific Science, 61(1), 3-16.
Henneman, M. L., & Memmott, J. (2001). Infiltration of a Hawaiian community by introduced biological control agents. Science, 293(5533), 1314-1316.
Hicks, D. J. (2015). Epistemological depth in a GM crops controversy. Studies in History and Philosophy of Science Part C: Studies in History and Philosophy of Biological and Biomedical Sciences, 50, 1-12.
Hill, T., Schlötterer, C., & Betancourt, A. J. (2016). Hybrid Dysgenesis in Drosophila simulans Associated with a Rapid Invasion of the P-Element. PLoS Genetics, 12(3), e1005920.
Hoffmann, A. A., Montgomery, B. L., Popovici, J., Iturbe-Ormaetxe, I., Johnson, P. H., Muzzi, F., ... & Cook, H. (2011). Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature, 476(7361), 454-457.
Hofschneider, A. (2016) Hawaii GMO Seed Companies Win Big In Appeals Court Rulings. Honolulu Civil Beat. Published online http://www.civilbeat.org/2016/11/hawaii-gmo-seed-companies-win-big-in-appeals-court-ruling/
Horn, C., & Wimmer, E. A. (2003). A transgene-based, embryo-specific lethality system for insect pest management. Nature Biotechnology, 21(1), 64-70.
Hourihan, M., & Parkes, D. (2016). Federal R&D in the FY 2016 Budget: An Overview. American Association for the Advancement of Science. Published online https://www.aaas.org/fy16budget/federal-rd-fy-2016-budget-overview
Howarth, F. G. (1983). Classical biocontrol: panacea or Pandora’s box. Proceedings of the Hawaiian Entomological Society, 24(2), 239-244.
IMF: International Monetary Fund (2016) World Economic Outlook Database. Published online http://www.imf.org/external/pubs/ft/weo/2016/01/weodata/index.aspx
Jayaraman, K. S., and C. Raghavan. 1975. The WHO and mosquitoes. Nature 257, 175-176.
Kahumoku, G. (1980). A Hawaiian Perspective on Taro Growing. Research Extension Series / Hawaiʻi Institue of Tropical Agriculture and Human Resources.
Kallis, T. 2013 Papaya: A GMO success story. Hawaii Tribune Herald, June 10. Available online http://hawaiitribune-herald.com/sections/news/local-news/papaya-gmo-success-story.html
Kathage, J., & Qaim, M. (2012). Economic impacts and impact dynamics of Bt (Bacillus thuringiensis) cotton in India. Proceedings of the National Academy of Sciences, 109(29), 11652-11656.
Kier, G., Kreft, H., Lee, T. M., Jetz, W., Ibisch, P. L., Nowicki, C., ... & Barthlott, W. (2009). A global assessment of endemism and species richness across island and mainland regions. Proceedings of the National Academy of Sciences, 106(23), 9322-9327.
Knipling, E. F., Laven, H., Craig, G. B., Pal, R., Kitzmiller, J. B., Smith, C. N., & Brown, A. W. A. 1968. Genetic control of insects of public health importance. Bulletin of the World Health Organization 38(3), 421.
Knols, B. G., Bossin, H. C., Mukabana, W. R., & Robinson, A. S. 2007. Transgenic mosquitoes and the fight against malaria: managing technology push in a turbulent GMO world. The American Journal of Tropical Medicine and Hygiene, 77(6 Suppl), 232-242.
Kolopack, P. A., Parsons, J. A., & Lavery, J. V. 2015. What makes community engagement effective?: lessons from the Eliminate Dengue Program in Queensland Australia. PLoS Neglected Tropical Disease 9(4), e0003713.
Langer, E. 1967. Chemical and Biological Warfare (I): The Research Program. Science 155(3759), 174-179.
Láruson, Á. J., & Reed, F. A. (2016). Stability of underdominant genetic polymorphisms in population networks. Journal of Theoretical Biology, 390, 156-163.
Laven, H. 1967. Eradication of Culex pipiens fatigans through cytoplasmic incompatibility. Nature 216, 383-384.
Laven, H., J. Cousserans, and G. Guille. 1972. Eradicating mosquitoes using translocations: a first field experiment. Nature 236, 456-457.
Liu, H. S., Jan, M. S., Chou, C. K., Chen, P. H., & Ke, N. J. (1999). Is green fluorescent protein toxic to the living cells? Biochemical and Biophysical Research Communications, 260(3), 712-717.
Loehle, C., & Eschenbach, W. (2012). Historical bird and terrestrial mammal extinction rates and causes. Diversity and Distributions, 18(1), 84-91.
Lynas, M. (2013). Time to call out the anti-GMO conspiracy theory. Mark Lynas speech hosted by the International Programs–College of Agriculture and Life Sciences (50th Anniversary Celebration), and the Atkinson Center for a Sustainable Future, Cornell University.
Lynas, M. (2013). The True Story About Who Destroyed a Genetically Modified Rice Crop. Future Tense: ASU | New America | Slate Published online http://www.slate.com/blogs/future_tense/2013/08/26/golden_rice_attack_in_philippines_anti_gmo_activists_lie_about_protest_and.html
Lyttle, T. W. (1981). Experimental population genetics of meiotic drive systems. III. Neutralization of sex-ratio distortion in Drosophila through sex-chromosome aneuploidy. Genetics, 98(2), 317-334.
Menon, P. (2001). Waking up to GM cotton. Frontline. 18(23). Available online http://www.frontline.in/navigation/?type=static&page=flonnet&rdurl=fl1823/18230440.htm
McGray, D. (2002). Biotech's Black Market. Mother Jones, September/October Issue. Available online http://www.motherjones.com/politics/2002/09/biotechs-black-market
Messing, R. H., & Wright, M. G. (2006). Biological control of invasive species: solution or pollution? Frontiers in Ecology and the Environment, 4(3), 132-140.
Mora, C., Frazier, A. G., Longman, R. J., Dacks, R. S., Walton, M. M., Tong, E. J., ... & Ambrosino, C. M. (2013). The projected timing of climate departure from recent variability. Nature, 502(7470), 183-187.
Morita, T., Kubota, H., Murata, K., Nozaki, M., Delarbre, C., Willison, K., ... & Gachelin, G. (1992). Evolution of the mouse t haplotype: recent and worldwide introgression to Mus musculus. Proceedings of the National Academy of Sciences, 89(15), 6851-6855.
Mutz, D. C. (1989). The influence of perceptions of media influence: Third person effects and the public expression of opinions. International Journal of Public Opinion Research, 1(1), 3-23.
Myers, N., Mittermeier, R. A., Mittermeier, C. G., Da Fonseca, G. A., & Kent, J. (2000). Biodiversity hotspots for conservation priorities. Nature, 403(6772), 853-858.
Nelson, S., Brooks, F., & Teves, G. (2011). Taro Leaf Blight in Hawaiʻi. College of Tropical Agriculture and Human Resources, University of Hawaiʻi, PD-71
Nemana, V. (2012). In India, Genetically Modified Crops Come at a High Price. The New York Times: India Ink. Published online http://india.blogs.nytimes.com/2012/10/16/in-india-gm-crops-come-at-a-high-price/
Nordlee, J. A., Taylor, S. L., Townsend, J. A., Thomas, L. A., & Bush, R. K. (1996). Identification of a Brazil-nut allergen in transgenic soybeans. New England Journal of Medicine, 334(11), 688-692.
NPR: National Public Radio (2013). Former Anti-GMO Activist Says Science Changed His Mind. All Things Considered, January 20, 2013. Available online http://www.npr.org/2013/01/20/169847199/former-anti-gmo-activist-says-science-changed-his-mind
Paine, Jacqueline A; Shipton, Catherine A; Chaggar, Sunandha; Howells, Rhian M; Kennedy, Mike J; Vernon, Gareth; Wright, Susan Y; Hinchliffe, Edward; Adams, Jessica L (2005). Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nature Biotechnology 23 (4): 482–7.
Pal, R. 1974. WHO/ICMR programme of genetic control of mosquitoes in India. In The Use of Genetics in Insect Control, Eds. R. Pal and M. J. Whitten. North-Holland, Amsterdam.
Parry, D. (2009). Beyond Pandora’s box: quantitatively evaluating non-target effects of parasitoids in classical biological control. Biological Invasions, 11(1), 47-58.
Perales, H. R., Benz, B. F., & Brush, S. B. (2005). Maize diversity and ethnolinguistic diversity in Chiapas, Mexico. Proceedings of the National Academy of Sciences of the United States of America, 102(3), 949-954.
Pimm, S. L., Russell, G. J., Gittleman, J. L., & Brooks, T. M. (1995). The future of biodiversity. Science, 269(5222), 347.
Powell K, Jayaraman K. S. 2002. Mosquito researchers deny plotting secret biowarfare test. Nature. 419: 867.
Potrykus, I. (2001) Golden Rice and Beyond. Plant Physiology 125 (3): 1157–1161.
Raghavan, C., and K. S. Jayaraman. 1975 The WHO and mosquitoes. Nature 257: 175-176.
Reeves, R. G., Denton, J. A., Santucci, F., Bryk, J., & Reed, F. A. (2012). Scientific standards and the regulation of genetically modified insects. PLoS Neglected Tropical Diseases, 6(1), e1502.
Reeves, R. G., Bryk, J., Altrock, P. M., Denton, J. A., & Reed, F. A. (2014). First steps towards underdominant genetic transformation of insect populations. PLoS One, 9(5), e97557.
Régnier, C., Bouchet, P., Hayes, K. A., Yeung, N. W., Christensen, C. C., Chung, D. J., ... & Cowie, R. H. (2015). Extinction in a hyperdiverse endemic Hawaiian land snail family and implications for the underestimation of invertebrate extinction. Conservation Biology, 29(6), 1715-1723.
Ritte, W., & Freese, B. (2006). Haloa. Seedling, (October), 11–14.
Sehgal NK. Doubts over US in India. Nature. 1974;251:177–178.
Serafino, N. M., Dale, C., Grimmett, R. F., Margesson, R., Rollins, J., Salaam-Blyther, T., Tarnoff, C., Woolf, A. F., Wyler, L. S., Bowman, S. (2008) CRS Report for Congress: The Department of Defense Role in Foreign Assistance: Background, Major Issues, and Options for Congress. Congressional Research Service; Foreign Affairs, Defense, and Trade Division. RL34639 Available online https://fpc.state.gov/documents/organization/110406.pdf
Séralini, G. E., Clair, E., Mesnage, R., Gress, S., Defarge, N., Malatesta, M., ... & De Vendômois, J. S. (2012). RETRACTED: Long term toxicity of a Roundup herbicide and a Roundup-tolerant genetically modified maize. Food and Chemical Toxicology, 50(11), 4221-4231.
Sharma, S. (2015) You should know: 5 deadliest mosquito-borne diseases in India. Hindustan Times. Available online http://www.hindustantimes.com/columns/you-should-know-5-deadliest-mosquito-borne-diseases-in-india/story-HVE7pprbScppKyqClE5GoN.html
Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N., Palmer, A. E., & Tsien, R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology, 22(12), 1567-1572.
Shaner, N. C., Steinbach, P. A., & Tsien, R. Y. (2005). A guide to choosing fluorescent proteins. Nature Methods, 2(12), 905-909.
Sinkins, S. P., & Gould, F. (2006). Gene drive systems for insect disease vectors. Nature Reviews Genetics, 7(6), 427-435.
SSWDC: Secretary of State Washington DC. (1975). STATE 248935: WHO genetic mosquito control project from Dr. Ehrlich, Director, HEW/OIH. Declassified/Released US Department of State EO Systematic Review. Retrieved from https://wikileaks.org/plusd/cables/1975STATE248935_b.html
Stone, G. D. (2010). The anthropology of genetically modified crops. Annual Review of Anthropology, 39, 381-400.
Tang, G; Qin, J; Dolnikowski, GG; Russell, RM; Grusak, MA (2009). Golden Rice is an effective source of vitamin A. Am J Clin Nutr. 89 (6): 1776–83.
Tomiche, F. J. 1975 The WHO and mosquitoes. Nature 257: 175.
Treaster, J. B. 1975 U.S. Health Service Aided Army on Poison Weapon. The New York Times. September 18, p. 1 col. 7, p. 26 col. 4.
Vitousek, P. M., (1988). Chapter 20: Diversity and Biological Invasions of Oceanic Islands. pp. 181-189. In Biodiversity. Eds. Wilson, E. O., Peter, F. M., National Academies Press, Washington, D.C.
Warner, R. E. (1968). The role of introduced diseases in the extinction of the endemic Hawaiian avifauna. The Condor, 70(2), 101-120.
Webber, B. L., Raghu, S., & Edwards, O. R. (2015). Opinion: Is CRISPR-based gene drive a biocontrol silver bullet or global conservation threat? Proceedings of the National Academy of Sciences, 112(34), 10565-10567.
Whitney, L. D., Bowers, F. A. I., & Takahashi, M. (1939). Taro Varieties in Hawaii. Hawaiʻi Agricultural Experiment Station of the University of Hawaiʻi. Bulletin 84.
Wimmer, E. A. (2005). Eco-friendly insect management. Nature Biotechnology, 23(4), 432-433.
Wimmer, E. A. (2013). Insect biotechnology: controllable replacement of disease vectors. Current Biology, 23(10), R453-R456.
Windbichler, N., Menichelli, M., Papathanos, P. A., Thyme, S. B., Li, H., Ulge, U. Y., ... & Crisanti, A. (2011). A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature, 473(7346), 212-215.
WHO: World Health Organization (1976) WHO-supported collaborative research projects in India: the facts. WHO Chronicle, 30: 131-139.
Xiao, Q. S., Xu, W. T., Yang, J. L., & Pan, L. W. (2016). Evaluation of genetically modified rice detection methods 2011/884/EU and 2008/289/EC proposed by the European Union. Journal of Integrative Agriculture, 15(12), 2899-2910.
Ye, X; Al-Babili, S; Klöti, A; Zhang, J; Lucca, P; Beyer, P; Potrykus, I (2000). Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 287 (5451): 303–5.
Zimmerman, H., Bloem, S., Klein, H., (2004) Biology, History, Threat, Surveillance and Control of the Cactus Moth, Cactoblastis cactorum. International Atomic Energy Agency, Austria. ISBN 92-0-108304-1. Available online: http://www-pub.iaea.org/mtcd/publications/pdf/faobsc_web.pdf