I sent my DNA sample to 23andme for SNP genotyping a little over a month ago. I just now received my results and my head is swimming from all the details. It is a bonanza of results to go over--I'm not even sure where to start. I plan to make a series of blog posts detailing some different aspects.
One service that they provide is the potential to contact and communicate with people who have matching chromosomal segments--i.e. relatives that share common ancestors. Of course I share half of my (autosomal) genome with each of my parents; and half of that (1/4) with each of my grandparents, 1/8 with my great grandparents, etc. The genome of an ancestor gets whittled away by recombination and the luck of transmission each generation down to us.
The expected size of the chromosome segment that gets passed on intact approximately follows a geometric distribution, which is a discrete form of the exponential distribution. One interpretation of this is the waiting time, distance, until an event, recombination, happens. The average length in Morgans is 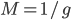 where
where 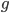 is the number of generations, because an exponential expectation is the inverse of the rate parameter (generations X recombination). The variance is
is the number of generations, because an exponential expectation is the inverse of the rate parameter (generations X recombination). The variance is 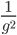 and the square-root of this gives us the standard deviation.
and the square-root of this gives us the standard deviation.
So in the graph above, after 10 generations we expect no recombination events within a chromosomal region with an average size of 10 cM. This can be thought of in either direction in time, the size of the region you pass on to descendants or the region you inherit from ancestors, or in both directions at once, back to a common ancestor and forward to a cousin--in this case 10 generations would be the distance to a 4th cousin. There is a wide variance, so 95% of the time you expect the identity-by-descent (IBD) tract to be less than 40 cM (plus two standard deviations). The lower bound on the interval size goes to zero and indeed, after a few generations we start loosing representation of ancestors in our genome (the number of ancestors grows exponentially, initially, and our genome is finite in size).
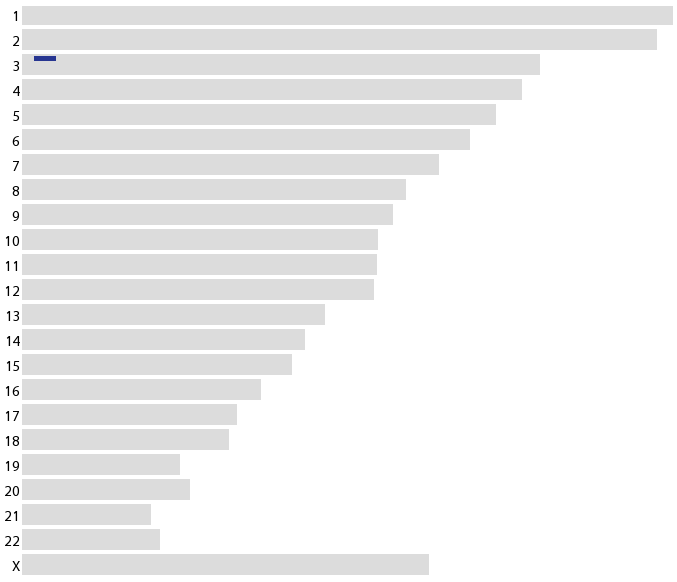
So, another individual, "M", that has also been genotyped by 23andme came up with some similarities and was flagged as a potential match. We contacted each other and found a shared 16.5 cM segment on chromosome 3 (the blue bar in the genome schematic below) consisting of over 2,000 genotyped SNPs (this is not just random chance).
This size segment is expected with six to twelve (+1 s.d.) generations between us. We compared genealogies and sure enough, we are descended from a family that lived in the 1800's in North Carolina. The parents of the family were Moses Pace (1781-1868) and Margaret Barclay (1793-1883). We are actually descended from two brothers that were their sons. William H. Pace (1826-1904) and Leander J. Pace (1816-1893). Here are pictures (this is the best image quality I have at the moment) of W. H. Pace (left) and L. J. Pace (right):
The pictures were made when the men were at different ages but they do look like they could be brothers. W. H. Pace is my g. g. great grandfather, five generations back. L. J. Pace is M's g. g. great grandfather, also five generations back. So we are 5th cousins separated by twelve generations, this is perfectly consistent with the expected size of the IBD.
Taking a step back for a moment and thinking about this, this result more or less proves the chain of ancestry back to these two brothers--through the intermediate ancestors. It is possible that the shared ancestry is from a different individual that we do not know about, but since we do have a paper trail and family tradition genealogy to this family, and the genetic results are consistent with the genealogical distance, it is a far simpler proposition to accept that this is indeed the relationship. Further matches with other descendants can help support or refute this.
The other interesting thing to realize that we have done here is reconstruct a bit of the genome, approximately 8 million base pairs of one chromosomal copy, of these two brothers (this can also help us phase the data but that is a different topic). We don't know exactly which parent the shared segment was inherited from, Moses Pace (1781-1868) or Margaret Barclay (1793-1883), but we do know (with the caveats above) that these brothers shared this segment. The more modern people that are genotyped and share their results the more we can reconstruct parts of ancestral genomes. This gives us genetic information that might be used to infer more about these individuals, not just predispositions to diseases but even things like possible personality traits and responses to stress. If the reconstruction is dense enough small gaps might be interpolated based on linkage and haplotype frequencies. It also might be possible to begin making shared ancestry links between reconstructed ancestral genomes to move even deeper into the past--this result has gone back about 200 years, another similar jump between ancestral relatives would put us back into the early 1600s! Finally, the parents of these brothers were born in the late 1700's; it is amazing to me that we can learn more about this family by the patterns shared in our DNA today.