For today I want to show something from a side-project that Aki Laruson is involved in. The aim here is to generate some preliminary data for a later grant application to expand the project in the near future. Here are pictures of two flies we generated that are genetic mosaics. They have groups of cells throughout their bodies that are made up of two different genotypes at a single chromosomal position (usually we think of all cells in our bodies as being genetically identical to each other, although that is overly simplistic for the most part it is true). In most tissues the genetic difference in these flies does not result in a visible genotype but in the eyes we can see the different cells in the flies. There are groups of cells that are either red or white phenotypes depending on their underlying genetic make-up. (Normally a Drosophila fruit fly would either have all red or all white cells in its eyes depending on what alleles it inherited from its parents.)
This was done using genetic sequences from a yeast plasmid (the two micron plasmid or 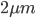 plasmid) that are inserted into the fly's genome (pioneered by Golic and Lindquist 1989 (PubMed, Google Scholar)). There is an enzyme called flippase (FLP) that recognizes (binds to) and cuts a specific sequence known as the Flippase Recognition Target (FRT), 5'-GAAGTTCCTATTCtctagaaaGtATAGGAACTTC-3'. In yeast the FLP-FRT system causes rearrangements to the plasmids structure that allow it to amplify in numbers in the cell when the cell undergoes DNA replication (e.g., Chan et al. 2013 (PubMed, Google Scholar)).
plasmid) that are inserted into the fly's genome (pioneered by Golic and Lindquist 1989 (PubMed, Google Scholar)). There is an enzyme called flippase (FLP) that recognizes (binds to) and cuts a specific sequence known as the Flippase Recognition Target (FRT), 5'-GAAGTTCCTATTCtctagaaaGtATAGGAACTTC-3'. In yeast the FLP-FRT system causes rearrangements to the plasmids structure that allow it to amplify in numbers in the cell when the cell undergoes DNA replication (e.g., Chan et al. 2013 (PubMed, Google Scholar)).
Here in flies the two FRT sites have been positioned on either side of part of the white gene sequence and FLP is placed under the Drosophila heat shock promoter (Hsp70). We exposed the larvae to 37 C temperatures (by immersing vials containing the larvae in warm water for an hour) to induce the expression of FLP by the flies normal gene activation reaction to heat, which then recombined the two FRT sites together and caused inactivation of the white gene, by deleting part of the gene sequence that was between the FRT sites, resulting in a loss of pigment in the cells of the eyes that are descended from cells in which this deletion happened in the larvae. (By the way, a common misconception is that the white gene encodes an eye pigment gene. It does not, but it is in a pathway that is required for the eventual production of several eye pigments in addition to other important molecules.) There are two broad categories of cells that we are interested in here, somatic and germline cells. Only germline cells produce gametes that result in the next generation so only the DNA from germline cells get passed on to the next generation; however, most of the cells in a multicellular animal's body are somatic cells that contain DNA that does not get passed on to the next generation. Ultimately we want flies that have the DNA rearrangement in all the cells of their body. We can see that the FLP-FRT rearrangement worked in some cells by the mosaic pattern in the eyes but we do not know if this has affected the germline of each individual fly. If it has, then any offspring arising from these affected cells will contain the genetic rearrangement in all of their cells (all of the cells will be descended from the same two starting gametes, genetic mosaics of this type should not be able to be passed on from generation to generation). Now we will breed these mosaic flies together and select for all white eyed flies that contain the deletion in all the cells of their body. Then it's on to the next stage of the experiment.

