I've been brushing up on DNA extraction, PCR (DNA amplification) and prepping the product for DNA sequencing (Sanger sequencing). The sequencing is done at a site on campus (link). We submitted our first DNAs Friday afternoon and got the results back by email Saturday afternoon! We tested a range of samples with a range of primers to see what worked and what didn't. (Part of this is getting ready for a genetics class I am teaching in the fall where I want students to sequence DNA from biological samples they collect.) My daughter found a praying mantis (Tenodera sinensis) in a parking lot so I snipped off a tiny piece of the end of one of its feet for a DNA sample.
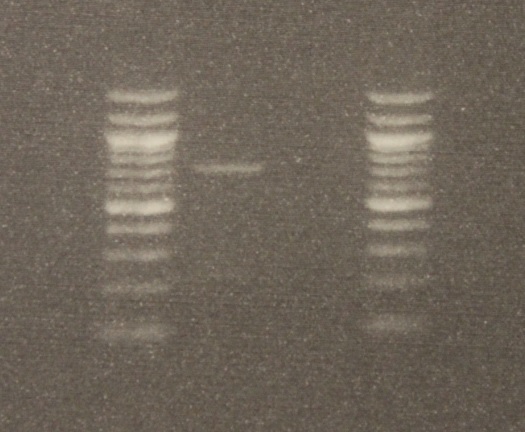
After DNA extraction I used two primers (short nucleotide sequences) to target a section of DNA, 710 base-pairs long (DNA letters), for amplification. (The primers are "LCO1490" 5'-GGTCAACAAATCATAAAGATATTGG-3' and "HCO2198" 5'-TAAACTTCAGGGTGACCAAAAAATCA-3', Folmer et al. 1994) The PCR worked! There was a band on the gel the right size. Below is an image of the DNA.
The gel image is read bottom up in columns. The first and last columns on the right and left with all the bright bands are DNA "ladders." Those are a collection of different sized DNAs to use as a reference. The bottom of the gel is positively charged and the top negative when it is running (before taking the image). This is done by hooking it up to 90 volts for half an hour. DNA is negatively charged so it runs down the gel. Smaller segments run faster than large ones so it separates a mixture by size. The PCR from the mantis sample is in the second row and a blank sample containing no DNA is beside it in the third row (this is a negative control for possible DNA contamination). By comparing the band from the mantis sample to the ladder we can see that a sequence between 700 to 800 base-pairs was amplified.
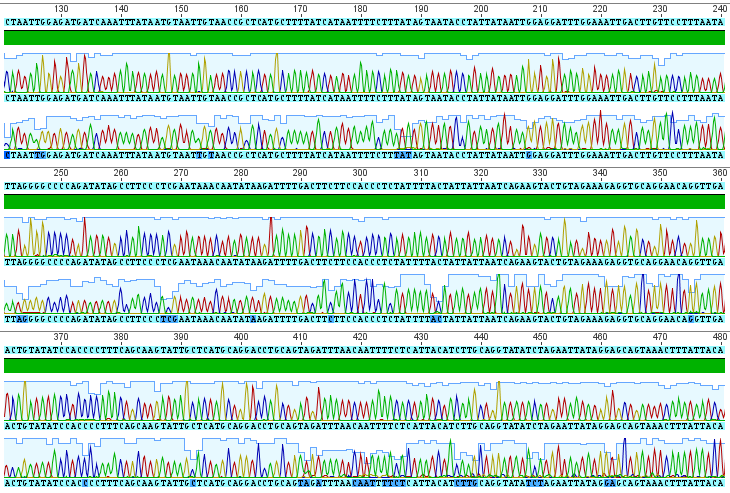
The sequences from this PCR amplification came back very nice with high quality signal. Here is a screen capture showing part of the "trace" file.
Each peak corresponds to a signal from a base-pair at that site along the DNA sequence (red=A, blue=C, yellow=G, green=T). There are two rows at each site because I sequenced both strands. The region I amplified is a small part of the cells mitochondrial genome, in a gene called COI for short (Cytochrome c Oxidase subunit I). From the results I get the following, cleaned up, 684 basepair DNA sequence (in fasta format).
>Mantis_COI
CATAAAGATATTGGAACACTATATTTTATTTTTGGTGCATGAGCAGGTATATTAGGAACATCTTTAAG
AATTCTAATTCGAACCGAATTAGGTCAACCAGGTTCCCTAATTGGAGATGATCAAATTTATAATGTAA
TTGTAACCGCTCATGCTTTTATCATAATTTTCTTTATAGTAATACCTATTATAATTGGAGGATTTGGAA
ATTGACTTGTTCCTTTAATATTAGGGGCCCCAGATATAGCCTTCCCTCGAATAAACAATATAAGATTT
TGACTTCTTCCACCCTCTATTTTACTATTATTAATCAGAAGTACTGTAGAAAGAGGTGCAGGAACAG
GTTGAACTGTATATCCACCCCTTTCAGCAAGTATTGCTCATGCAGGACCTGCAGTAGATTTAACAAT
TTTCTCATTACATCTTGCAGGTATATCTAGAATTATAGGAGCAGTAAACTTTATTACAACTATAATTAAT
ATAAAACCATTATATATAAATCAAACTCAAGTTCCCCTTTTTGTTTGATCCGTTGGTATTACAGCTTTA
TTACTTCTATTATCATTACCTGTTCTTGCAGGAGCAATTACTATATTATTAACTGATCGAAATCTAAATA
CCTCATTTTTTGATCCTGCTGGAGGAGGTGATCCTATTCTTTATCAACACTTATTTTGATTTTTTGGT
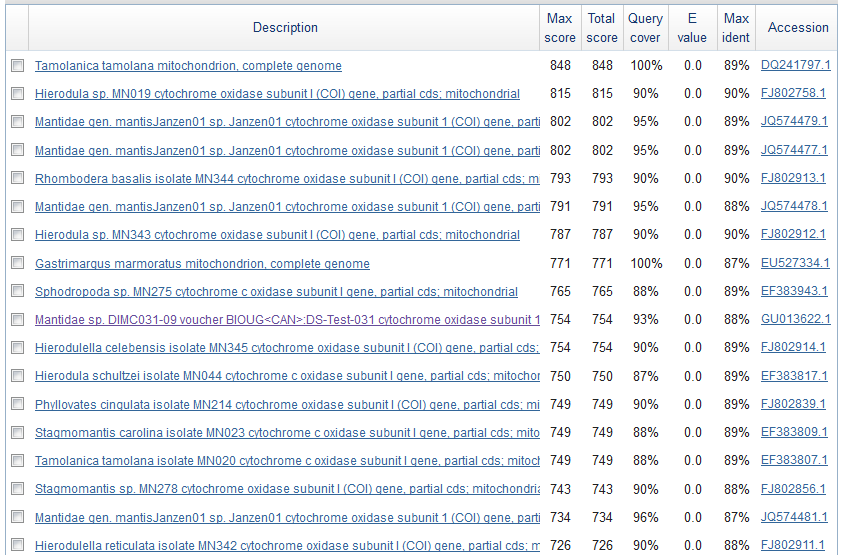
Just to check if this is the right sequence (I might have accidentally switched samples or there is always a possibility of DNA contamination) I searched for similar sequences using BLASTn on GenBank. Here is a screen shot of the most similar sequences.
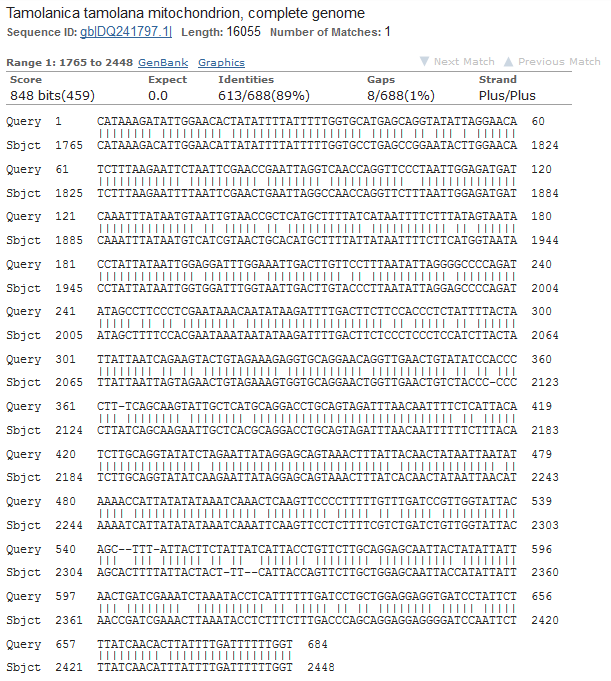
These are Mantis (Mantidae family) species that are returned so this looks correct. The top match is compared below.
Tamolanica tamolana, the shield mantis from New Guinea, is 89% identical.