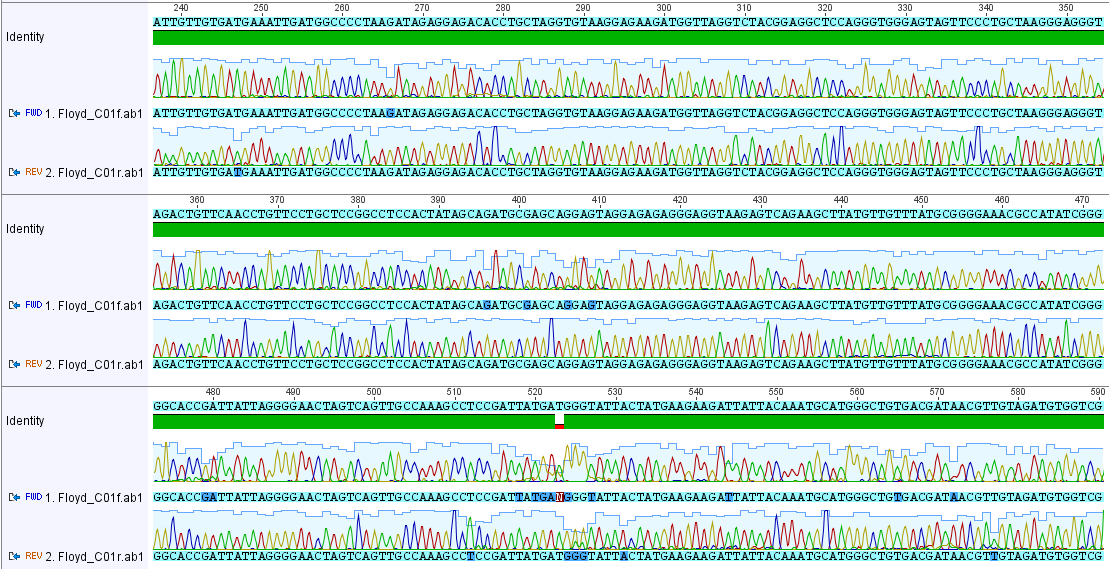
I have been trying different methods to isolate DNA from cheek cells. I tried a "BuccalAmp" kit on a recommendation and it works really well. It is quick and easy. I tried the COI barcode PCR on my own DNA sample and compared the results to the earlier DNA extractions and PCR from three species of insects. First here is part of the aligned traces from forward and reverse sequencing of the amplified product.
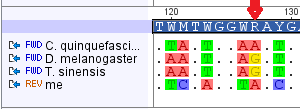
And here is an alignment with the fruitfly, mantis and mosquito sequences (the basepairs that are different are highlighted).
You can probably see from the alignment that a lot of the changes are with my sequence and that the insects are closer related to each other. There are a few in-congruent homoplasies however. Here is one at position 127 in the alignment.
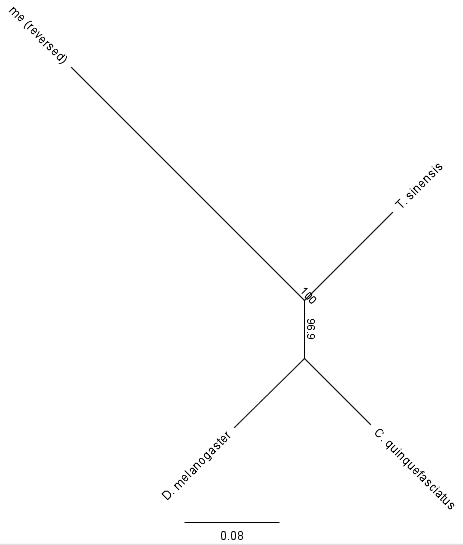
Humans and mosquitoes share an "A" at this position while fruit-flies and mantises share a "G," recall that mosquitoes are closer related to fruit-flies than to mantises. This is just an example of mutations that have occurred more than once at the same position. Making a "tree" to represent the mutations that are shared and different gives this pattern:
There is a long distance between me and the insects, as expected. Additionally, the mosquito (C. quinquefasciatus) groups closer to the fruit-fly (D. melanogaster) than to the mantis (T. sinensis). The number in the middle is the bootstrap support value for the tree. In bootstrapping the DNA bases are randomly shuffled and sampled with replacement to make new sequences the same length as the original (and by chance some basepairs are left out while others can be included more than once) the amount of time the grouping of species below that node in the tree is found is indicated. So, mosquitoes and fruit-flies are grouped together 96.9% of the time with random shuffling of their sequences (rather than one of these being closer to the mantis), which indicates a high amount of support form the data-set that this is the actual evolutionary pattern (with assumptions...).
This mitochondrial sequence is inherited from my mother, and her mother, on back in a maternal linage, changing only occasionally by mutations. My brother and sister share the same sequence as myself but I will not pass it on to my children, because the mitochondria is not transmitted from the father, only from the mother. I compared my sequences to others on genbank and found something funny.
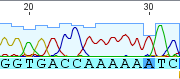
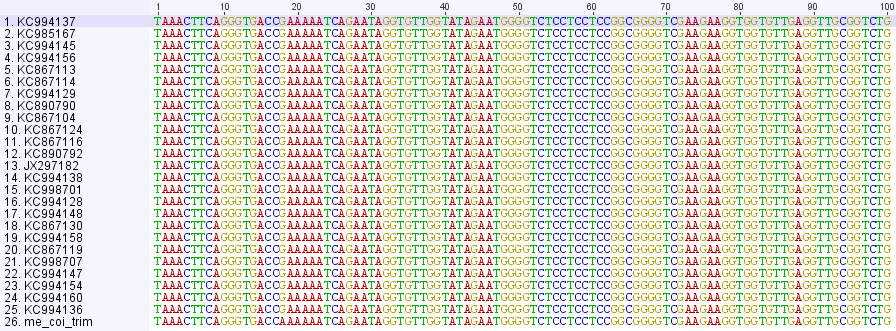
Near the beginning of the sequence I have an "A" at position 25 (above) that begins a string of six A's. However none of the other human mitochondrial sequences on genbank share this. Below is a comparison of 25 of the most similar human mitochondrial sequences from genbank to my sequence at the end, they are all identical with me except at this position (at position 18 in the alignment below):
They all have a G followed by five A's. Since this seems to be rare, it might be interesting to see if anyone out there has the same sequence. I can not go back very far, only about four generations, along my maternal lineage. My matrilineal Great Grandmother's maiden name was Sarah J. Carlisle born in 1873 in North Carolina; her mother was Dicey (which might be short for Leodicia) Owenby (or Owensby), born about 1834 in North Carolina, and her mother might have been Sarah Hunter born around 1815 in South Carolina--just in case anyone that might be related reads this.