I've added a few more species to our plant chloroplast rbcL DNA sequence collection.
The first two new ones, above and below, I bought at the local store: sweet corn (Zea mays) and papaya (Carica papaya).
There is also a type of flower blooming all around campus this time of year (March-April). They range from white to dark purple. I am guessing that they might be a good example of incomplete dominance (in genetics classes we often use white/pink/red snapdragons as an example, but these may be a nice local example that students actually see outside of class). I looked them up and they are Chinese Violets (Asystasia gangetica).
Above, purple on the right, to light purple in the middle, to white on the left.
The plant below (it looks like orange threads) is very interesting. It is called western field dodder (Cuscuta campestris) and is a parasite that attaches to and takes nutrients from other plants. Most plants use their chloroplasts to produce energy, but the dodder has come up with an alternative strategy. So, what does its chloroplast sequence look like?
Some places online say that the dodder does not have chloroplasts or produce chlorophyll. However, this is not true. Funk et al. (2007) sequenced the chloroplast genome of two dodder species (C. reflexa and C. gronovii) and found that they had reduced and rearranged genomes. McNeal et al. (2007) also found some gene loss and rearrangements, and an increase in the nucleotide substitution rate, in two other dodders (C. exaltata and C. Obtusiflora). Furthermore, Berg et al. (2004) found that C. gronovii and C. subinclusa have lost the plastid RNA polymerase (necessary for gene expression by generating a messenger RNA, i.e. transcription) and that rbcL has had to evolve to be transcribed by RNA polymerase from the nuclear genome.
Below is the DNA sequence alignment from all eight species (including the four from the earlier post). There are nine entries because I included two Chinese violets, a white form and a purple form, and unsurprisingly they have identical sequences.
I changed the settings so that any DNA position that is variable is highlighted and positions that are the same in all species are represented by a ".", except in the consensus sequence given along the top. Overall, you may notice a 2-1-2-1-2-1 pattern in the spacing of variable sites. Below is a close up of a stretch where this is particularly strong.
In this part of the rbcL gene every three nucleotides codes for a particular amino acid. Below each DNA sequence I have listed the corresponding amino acid code. Across all eight species the amino acid sequence is identical (in this region; there are some variations in some other parts of the gene) corresponding to "TSIVGNVFGFKALRALRLEDLRIP." In other words, none of the DNA changes affect the sequence of the protein enzyme (enzymes catalyze biochemical reactions (catalyze = cause the reaction to occur)) that is produced by the gene. The sets of three nucleotides that code for an amino acid are known as codons. If you look at a codon table that translates between nucleotides and amino acids you can see that any change to the second position causes a change in the corresponding amino acid. However, often changes in the third position have no effect on the amino acid (i.e. they are "silent"). For example, GCA, GCC, GCG, and GCT all code for alanine (A). Occasionally changes to the first position do not change the amino acid; so both TTA and CTA code for leucine (L). This explains the spacing of the DNA sequence changes between these species. Mutations are expected to be happening across all of the sites. However, ones that change the protein sequence may affect the function of the enzyme and are removed by selection. So, over time, the only changes that are accumulating between species are ones that are not affected by selection (i.e. are selectively "neutral").
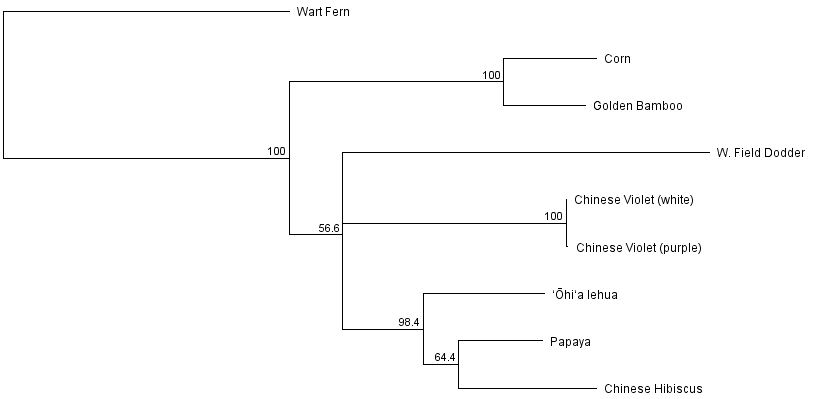
Below is a phylogenetic tree that can be obtained from these sequences to represent their inferred evolutionary history (rooted by the fern sequence as an outgroup).
Corn and bamboo are both types of grasses so it makes sense that they group together. The dodder has a long branch, representing many DNA changes, which fits with what we expect given the extensive changes and higher rates of substitutions in the dodder chloroplast genome. The two Chinese violets cluster together identically, which makes sense because they are the same species. There could be a small amount of genetic variation within the species between the two samples but it is also not surprising that there is not in this particular sequence. The remaining branches are the lehua, hibiscus and papaya which are all considered Rosids within the Eudicots (true dicots) while the dodder and Chinese violet are considered Asterids within the Eudicots.
So how much confidence do we have in the branching pattern in this tree? One way to address this is by "bootstrapping." This is a process where a large number of fake samples are generated by randomly sampling from the original dataset (randomly picking nucleotide sites) and the percent of time a particular branch of the tree is found (contains the same set of descentants) is used as a measure of confidence in that part of the tree. However, if many different branching orders are found, and different groups of descendants are contained within the branch, from different sets of the data then we have lower confidence in that particular feature. (See Felsenstein (1985) and Efron et al. (1996) for more information.) In the tree below I have applied bootstrapping and only shown the branches that are supported in a majority of the replicates. If there is no majority (e.g. if three different branching orders are found at equal 1/3 frequency) that part of the tree is collapsed together.
Several parts of the tree have high support from the data and are recovered >95% of the time (in fact the grass grouping was found 100% of the time). But the papaya hibiscus grouping had low support (64.4%) and branching order at the base of the eudicots (asterids and rosids) have little support in this dataset and so at the moment should be taken with a grain of salt (i.e. without confidence).